Team:Caltech/Week 6
From 2010.igem.org
(Difference between revisions)
(updated wed-sun) |
|||
| Line 18: | Line 18: | ||
** Plated cells every hour for three hours | ** Plated cells every hour for three hours | ||
** Combined glycerol solutions with LB and plated the solution | ** Combined glycerol solutions with LB and plated the solution | ||
| - | + | * New 16hr ligation procedure: | |
| + | ** 30μL total: 2μL T4 ligase, 3μL 10x ligase buffer, 5μL vector, 10μL of each brick. Incubate for 16hr at 16°C. | ||
| + | * Started more cultures to miniprep tomorrow: I13504, J04450, K274002, M30109. | ||
==Wednesday 7/21== | ==Wednesday 7/21== | ||
| - | * | + | * Transformed 3μL of each ligation product from yesterday (time constants all w/in 3.8-4.0). Plated 50μL of each on plates. |
==Thursday 7/22== | ==Thursday 7/22== | ||
| + | * Prepared new sequencing primers in preparation for CPCR. | ||
| + | * Transformation plates are almost completely empty - will centrifuge and plate remaining transformation cells. | ||
| + | * Colony PCR mixture: | ||
| + | ** 10μL 10x PCR buffer, 2μL dNTP mix, 1μL forward primer, 1μL reverse primer, 0.5μL Taq polymerase, fill to 100μL total. (Optional: add 20μL Qiagen Q buffer. Doesn't appear to help any of our reactions.) | ||
| + | ** Pick colony and add to water (85.5μL). Incubate at 100°C for 10min. Add remaining ingredients and begin PCR program: | ||
| + | *** Initial denaturation: 94°C, 4min | ||
| + | *** 30 cycles: | ||
| + | **** 94°C, 30s | ||
| + | **** 54°C, 30s | ||
| + | **** 72°C, 2min (1min/kb) | ||
| + | *** Final extension: 72°C, 10min | ||
| + | *** Finish: 5°C forever | ||
| + | * Ran colony PCR on 4 ligations, 2 had the correct insert length: L17, L27. | ||
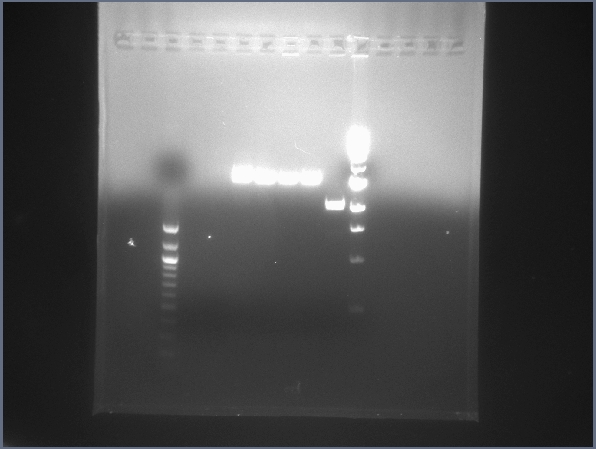
* Ran gel of digest products from yesterday to verify correctness of the digestion. | * Ran gel of digest products from yesterday to verify correctness of the digestion. | ||
[[Image:Digest22-07.jpg|200px|thumb|right|Digest test gel with upstream, downstream, and backbone digested bricks. (7/22)]] | [[Image:Digest22-07.jpg|200px|thumb|right|Digest test gel with upstream, downstream, and backbone digested bricks. (7/22)]] | ||
==Friday 7/23== | ==Friday 7/23== | ||
| + | * Plates containing the centrifuged transformation cells contained many colonies! | ||
| + | * Began 21 more CPCR reactions on ligations: L17, L18, L21, L27, L29, L16, L26, L25 | ||
| + | ** Note that because L18 is a coding sequence, it required the coding prefix primer; all others used the noncoding prefix primer. | ||
| + | * Received our heat shock promoter from the Registry today! Made a plate and culture of this brick (K112400). | ||
| + | * Purified yesterday's 4 CPCR reactions using a PCR purification kit. | ||
| + | ** Concentrations of each are all quite low (<20ng/μL) - we may want to increase the number of PCR cycles. | ||
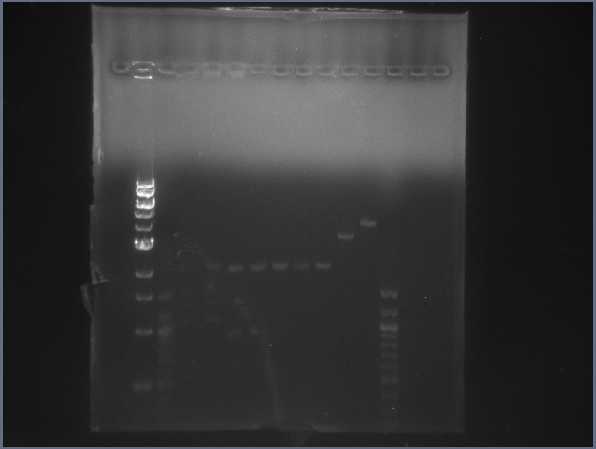
| + | * Ran 2 gels of all the CPCR results: | ||
| + | |||
==Weekend 7/24-25== | ==Weekend 7/24-25== | ||
| + | === Saturday 7/24=== | ||
| + | * Started 4 new CPCR reactions on L21 and L26 to verify them more accurately. | ||
| + | ** Disposed of all failed cultures... | ||
| + | ** Prepped successful clones: 17-6, 18-3, 21-3, 26-1, 27-4, 29-2, and the heat shock promoter (K112400). | ||
| + | * Prepared digest: | ||
| + | ** K215000 (u/d) | ||
| + | ** K112400 (u/d) | ||
| + | ** B0034 (u) | ||
| + | ** R0082 (u) | ||
| + | ** L17 (u) | ||
| + | ** L18 (u/d) | ||
| + | ** L26 (u) | ||
| + | ** I13504 (d) | ||
| + | ** L21 (d) | ||
| + | * Began ligations: L16, L19, L42, L43, L44, L23, L24, L31, L40, L34, L35 | ||
| + | * Ran another gel of the CPCR products: | ||
| + | |||
| + | === Sunday 7/25 === | ||
| + | * Transformed all ligations from yesterday (time constants all within 3.6-4.0). Used 4μL for all, except for L34 & L35, which sparked, so used 3μL instead. | ||
| + | * Made 2YT for making more electrocompetent cells. | ||
| + | * Plated all transformation cells (centrifuged at 1000g for 15min). | ||
| + | |||
| | ||
}} | }} | ||
Revision as of 04:43, 28 July 2010
|
People
|
Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 | Week 8 | Week 9 | Week 10 | Week 11 | Week 12 | Week 13 | Week 14 | Week 15
Monday 7/19
Tuesday 7/20
Wednesday 7/21
Thursday 7/22
Friday 7/23
Weekend 7/24-25Saturday 7/24
Sunday 7/25
|
 "
"