Team:Bielefeld-Germany/Results/Characterization
From 2010.igem.org
(Difference between revisions)
(→Tested BioBricks) |
|||
| Line 9: | Line 9: | ||
| - | ===<partinfo>BBa_K238011</partinfo>: ''vir''- | + | ===<partinfo>BBa_K238011</partinfo>: ''vir''-promoter=== |
We made a restriction analysis and sequenced parts of this BioBrick. | We made a restriction analysis and sequenced parts of this BioBrick. | ||
| Line 15: | Line 15: | ||
===<partinfo>P1010</partinfo>: ''ccdB''-gene=== | ===<partinfo>P1010</partinfo>: ''ccdB''-gene=== | ||
We transformed this BioBrick into ''Escherichia coli'' JM109, DH5α, TOP10 and DB3.1. ''E. coli'' JM109 and DH5α seem to be ''ccdB'' resistant because there were as much colonies after P1010 transformation as observed with DB3.1. The P1010 works as expected in ''E. coli'' TOP10 (no colonies after transformation) and DB3.1 (lawn after transformation). | We transformed this BioBrick into ''Escherichia coli'' JM109, DH5α, TOP10 and DB3.1. ''E. coli'' JM109 and DH5α seem to be ''ccdB'' resistant because there were as much colonies after P1010 transformation as observed with DB3.1. The P1010 works as expected in ''E. coli'' TOP10 (no colonies after transformation) and DB3.1 (lawn after transformation). | ||
| + | |||
| + | |||
| + | {|cellpadding="10" style="border-collapse: collapse; border-width: 1px; border-style: solid; border-color: #000" | ||
| + | |- | ||
| + | !style="border-style: solid; border-width: 1px"| ''E. coli'' strain | ||
| + | !style="border-style: solid; border-width: 1px"| Resistant to <partinfo>P1010</partinfo>? | ||
| + | !style="border-style: solid; border-width: 1px"| Expected result? | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| DB3.1 | ||
| + | |style="border-style: solid; border-width: 1px"| + | ||
| + | |style="border-style: solid; border-width: 1px"| + | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| DH5α | ||
| + | |style="border-style: solid; border-width: 1px"| + | ||
| + | |style="border-style: solid; border-width: 1px"| - | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| JM109 | ||
| + | |style="border-style: solid; border-width: 1px"| + | ||
| + | |style="border-style: solid; border-width: 1px"| - | ||
| + | |- | ||
| + | |style="border-style: solid; border-width: 1px"| TOP10 | ||
| + | |style="border-style: solid; border-width: 1px"| - | ||
| + | |style="border-style: solid; border-width: 1px"| + | ||
| + | |- | ||
| + | |} | ||
| + | |||
===<partinfo>K389011</partinfo>: VirA screening device=== | ===<partinfo>K389011</partinfo>: VirA screening device=== | ||
[[Image:IGEM-Bielefeld-LD50.jpg|600px|thumb|center|Ratio of surviving colonies of ''E. coli'' EC100D carrying unmutated <partinfo>K389010</partinfo> and <partinfo>K389011</partinfo> plated on PA agar plates with chloramphenicol, ampicillin and different concentrations of kanamycin. Comparison between cells that were induced with acetosyringone with cells that were not induced.]] | [[Image:IGEM-Bielefeld-LD50.jpg|600px|thumb|center|Ratio of surviving colonies of ''E. coli'' EC100D carrying unmutated <partinfo>K389010</partinfo> and <partinfo>K389011</partinfo> plated on PA agar plates with chloramphenicol, ampicillin and different concentrations of kanamycin. Comparison between cells that were induced with acetosyringone with cells that were not induced.]] | ||
| + | |||
=Sequenced BioBricks= | =Sequenced BioBricks= | ||
| + | |||
===Own BioBricks=== | ===Own BioBricks=== | ||
| Line 48: | Line 76: | ||
* <partinfo>K389050</partinfo> | * <partinfo>K389050</partinfo> | ||
| + | |||
===Other BioBricks=== | ===Other BioBricks=== | ||
Revision as of 18:11, 9 October 2010
{{{1}}}
Contents |
Tested BioBricks
We tested the following BioBricks:
<partinfo>K238008</partinfo>: virA
We made a restriction analysis and sequenced parts of this BioBrick. There were problems - more information within a short time.
<partinfo>BBa_K238011</partinfo>: vir-promoter
We made a restriction analysis and sequenced parts of this BioBrick.
<partinfo>P1010</partinfo>: ccdB-gene
We transformed this BioBrick into Escherichia coli JM109, DH5α, TOP10 and DB3.1. E. coli JM109 and DH5α seem to be ccdB resistant because there were as much colonies after P1010 transformation as observed with DB3.1. The P1010 works as expected in E. coli TOP10 (no colonies after transformation) and DB3.1 (lawn after transformation).
| E. coli strain | Resistant to <partinfo>P1010</partinfo>? | Expected result? |
|---|---|---|
| DB3.1 | + | + |
| DH5α | + | - |
| JM109 | + | - |
| TOP10 | - | + |
<partinfo>K389011</partinfo>: VirA screening device
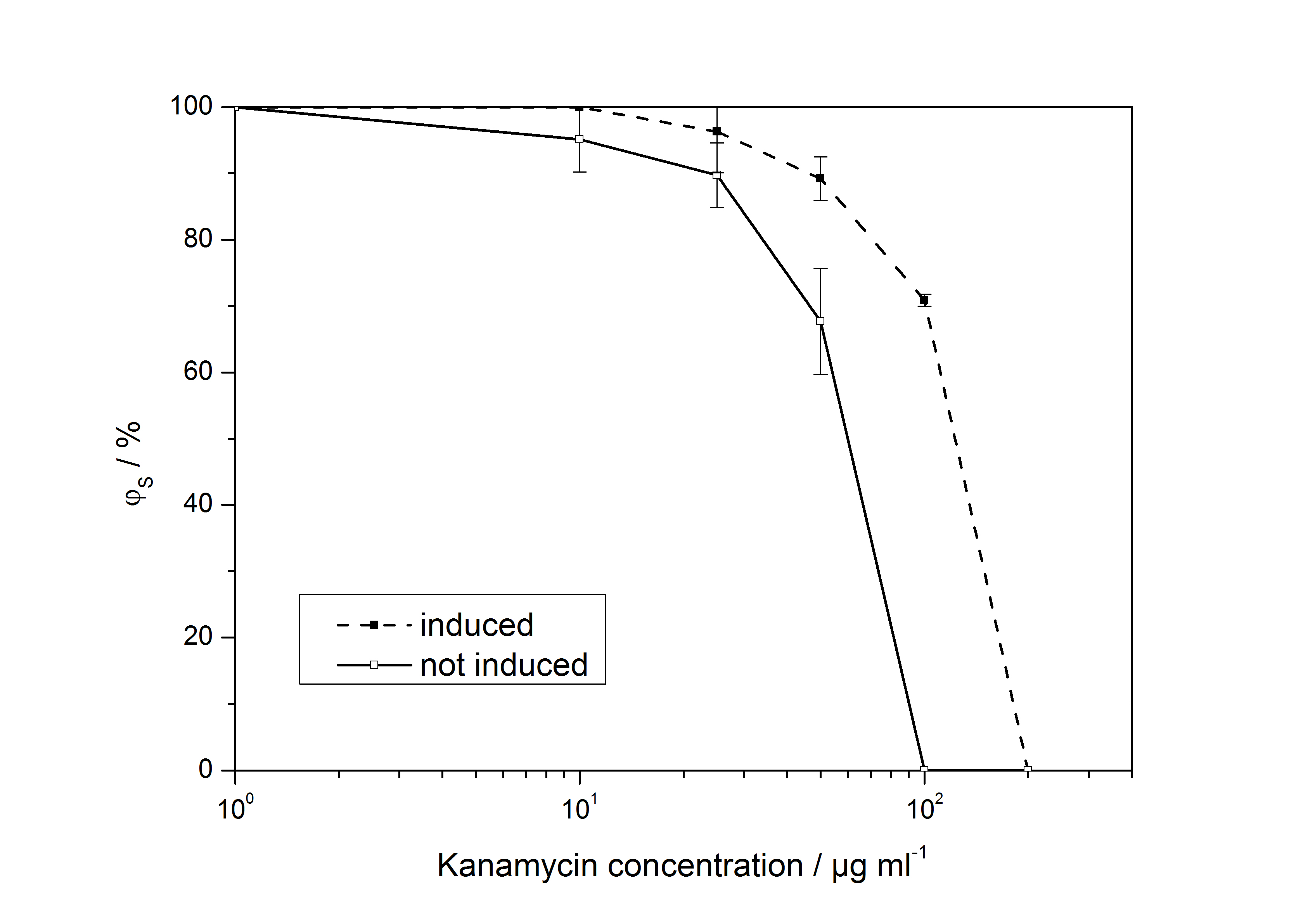
Ratio of surviving colonies of E. coli EC100D carrying unmutated <partinfo>K389010</partinfo> and <partinfo>K389011</partinfo> plated on PA agar plates with chloramphenicol, ampicillin and different concentrations of kanamycin. Comparison between cells that were induced with acetosyringone with cells that were not induced.
Sequenced BioBricks
Own BioBricks
The sequencing of the following of our own BioBricks was succesful and lead to the expected results:
- <partinfo>K389001</partinfo> (not fully completed)
- <partinfo>K389002</partinfo>
- <partinfo>K389003</partinfo>
- <partinfo>K389004</partinfo>
- <partinfo>K389005</partinfo>
- <partinfo>K389010</partinfo> (not fully completed)
- <partinfo>K389011</partinfo> (not fully completed)
- <partinfo>K389012</partinfo> (not fully completed)
- <partinfo>K389013</partinfo> (not fully completed)
- <partinfo>K389015</partinfo> (not fully completed)
- <partinfo>K389016</partinfo> (not fully completed)
- <partinfo>K389050</partinfo>
Other BioBricks
- <partinfo>K238008</partinfo> sequencing gave negative results - infos in registry are not correct!
- <partinfo>K238011</partinfo> sequencing gave negative results - infos in registry are not correct!
 "
"


