Team:Bielefeld-Germany/Project/Approach
From 2010.igem.org
| Line 48: | Line 48: | ||
We use high amounts of the antibiotic kanamycin in order to select the most specific system for our substances. By editing the degree of kanamycin we are able to carefully select the best mutant out of our PCR-tests. The mutated ''virA'' system will be induced after the error prone PCR by a mix of possible targets for the system (like capcaicin, dopamin, homovanillic acid etc.). It is important to avoid any acetosyringone, so we will be able to search for systems with new targets. | We use high amounts of the antibiotic kanamycin in order to select the most specific system for our substances. By editing the degree of kanamycin we are able to carefully select the best mutant out of our PCR-tests. The mutated ''virA'' system will be induced after the error prone PCR by a mix of possible targets for the system (like capcaicin, dopamin, homovanillic acid etc.). It is important to avoid any acetosyringone, so we will be able to search for systems with new targets. | ||
| - | If we do not handle with a lack of time, we are going to transform a sensitivity tuner into the BioBricks (compare Cambridge 2009). This will increase the sensitivity of our system. | + | |
| + | === The final construct=== | ||
| + | |||
| + | If we do not handle with a lack of time, we are going to transform a sensitivity tuner into the BioBricks (compare Cambridge 2009). This will increase the sensitivity of our system. So our final system will work like this: | ||
| + | |||
| + | [[Image:Bielefeld_Vorgehen.gif]] | ||
Revision as of 11:35, 27 August 2010
Contents |
The Approach
First we looked for a sensor system which is able to detect substances of interest. Moreover we needed substances of interest.The system of interest has to be taken out of a differnent bacteria species than Escherichia coli , in order to avoid any background. Therefore we checked the literature for a well reviewed sensor system, which is not part of the E. coli genome. The system of choice is the phenolic sensing system virA of Aggrobacterium tumefaciens. The systems naturally detects acetosyringone, which is a secondary metabolite of plants that affects bacteria as an attractant. So we hit the literature again to check for any other possible substances, which could be detected by the system. We got a really long list of possibilities and picked 'capcaicin', which is responsible for the spiciness of a fare.
The biological steps for creating a new sensor system are:
- extracting the virA system out of A. tumefaciens
- create new BioBricks out of the exciting environmental parts
- transform the new BioBricks into E. coli
- modify the system for sensibility and specificity by error prone PCR
- find and select the most promising mutants
Preparing the system
We tried to work with the already existing virA out of the registry (<partinfo>K238008</partinfo>). Unfortunately this virA BioBrick did not work. The sequence data of the BioBrick did not fit to the published virA sequence. So we had to extract the virA gene via PCR out of the A. tumefaciens TI-plasmid by ourselves in order to create a new BioBrick.
VirA does not work without the help of the VirG protein. The existing virG gene in the iGEM registry contains illegal restriction sites. Moreover it needs the help of Chev Protein and sugars to work perfectly with VirA. We decided to get some work done by Mr. Gene. So we changed the sequence manually and get the gene synthesized.
Starting point for biobricks -> Go cloning
The applied screening system in E. coli consists of two plasmids. After discussing the possibility of creating only one big plasmid, we unify that one plasmid will minimize the transformation efficiency and will be difficult to modify via error prone pcr. So we had to change the origin of a pSB1X3 plasmid in order to avoid compatibility problems at the transformation. Therefore we cloned the R6K origin into the <partinfo>pSB1A3</partinfo>, <partinfo>pSB1C3</partinfo> and <partinfo>pSB1T3</partinfo> plasmids. For this purpose we used the R6K sequence <partinfo>J61001</partinfo> and the pSBXXX sequence to create primers for the PCR following the ligation of the PCR products.
The error prone PCR
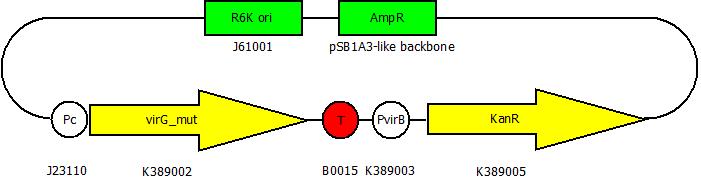
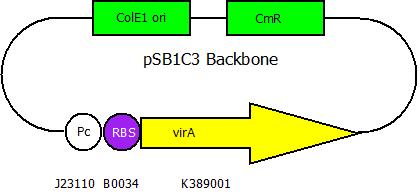
After cloning the new origin into the pSBXXX-backbones we are able to create the two constructs. The first one will be inside the competent bacteria cells and contains the virG gene under the constitutive promotor <partinfo>J23110</partinfo>, a terminator (<partinfo>B0017</partinfo>), a vir promotor and a readout or selection gene (luciferase, mRFP and kanamycin resistance, respectively):
The second plasmid contains the virA gene under the control of the constitutive promotor <partinfo>J23110</partinfo> and will be transformed and modified in one step via error prone PCR.
The error prone PCR is a PCR under malfunction conditions for the polymerase. It is possible to regulate the frequency of the mutagenesis by editing unbalanced concentrations of nucleotides and co-factors to the PCR. So we are going to create a new BioBrick and transform it into our target by one step. Before we are able to do the error prone PCR we need to get the backbones done.
Survival of the fittest
We use high amounts of the antibiotic kanamycin in order to select the most specific system for our substances. By editing the degree of kanamycin we are able to carefully select the best mutant out of our PCR-tests. The mutated virA system will be induced after the error prone PCR by a mix of possible targets for the system (like capcaicin, dopamin, homovanillic acid etc.). It is important to avoid any acetosyringone, so we will be able to search for systems with new targets.
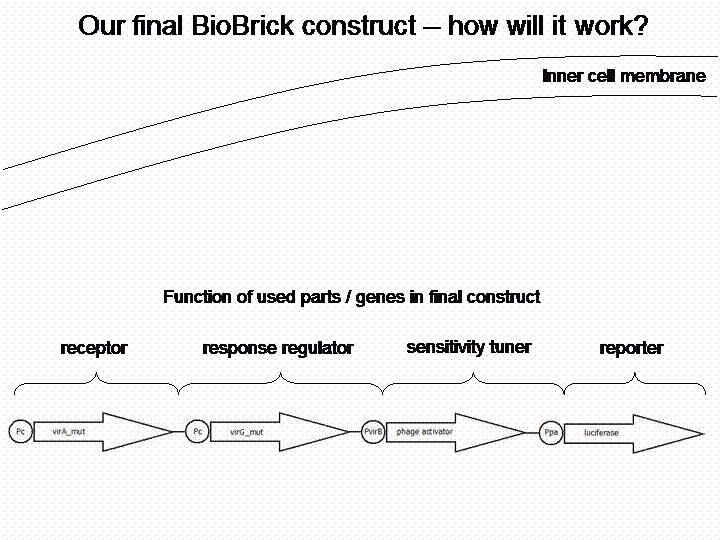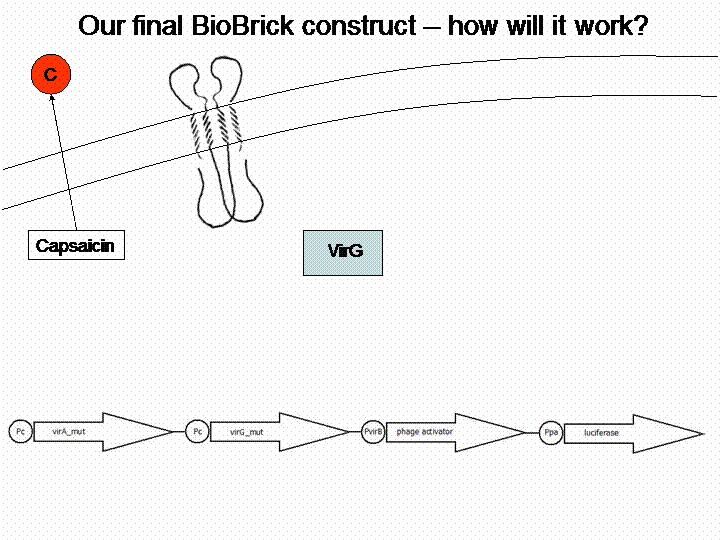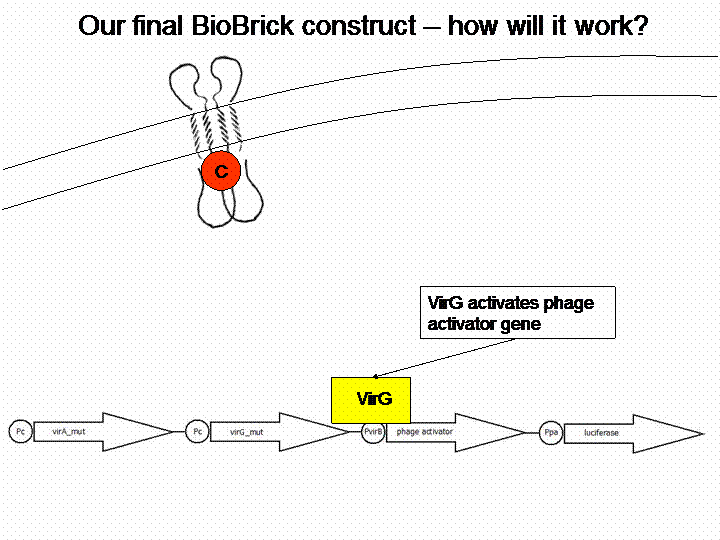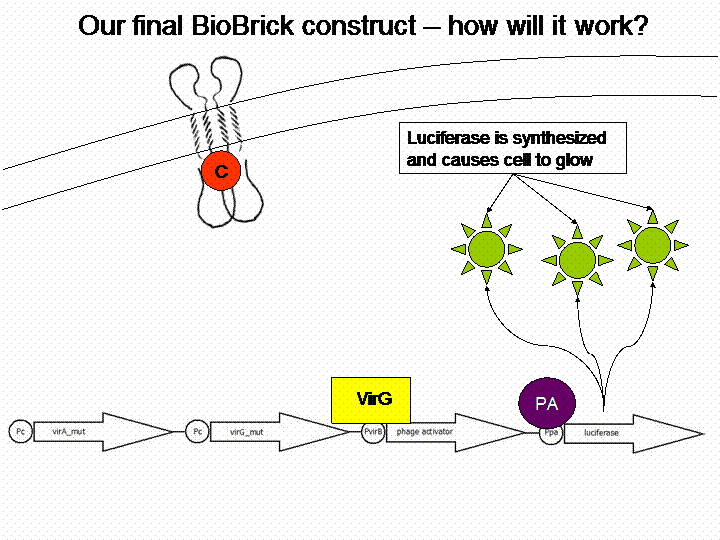
The final construct
If we do not handle with a lack of time, we are going to transform a sensitivity tuner into the BioBricks (compare Cambridge 2009). This will increase the sensitivity of our system. So our final system will work like this:
 "
"