Team:Bielefeld-Germany/Results/Submitted
From 2010.igem.org
(→BioBrick for virA-screenings) |
|||
| (14 intermediate revisions not shown) | |||
| Line 22: | Line 22: | ||
We have submitted the following [[Team:Bielefeld-Germany/Results/Characterization | working]] BioBricks. For a more detailed theoretical background to these BioBricks check our [[Team:Bielefeld-Germany/Project/Theory | theory pages]], too. | We have submitted the following [[Team:Bielefeld-Germany/Results/Characterization | working]] BioBricks. For a more detailed theoretical background to these BioBricks check our [[Team:Bielefeld-Germany/Project/Theory | theory pages]], too. | ||
| + | |||
===VirA receptor=== | ===VirA receptor=== | ||
| - | [[Image:Vira_rezeptor.jpg|300px|thumb|right|''The VirA receptor of ''Agrobacterium tumefaciens''.'']] | + | [[Image:Vira_rezeptor.jpg|300px|thumb|right|'''Fig. 1: The VirA receptor of ''Agrobacterium tumefaciens''.''']] |
| - | The VirA receptor is used by ''A. tumefaciens'' to detect acetosyringone and other phenolic substances which are secreted by plants after injury. In presence of these substances VirA phosphorylates itself and afterwards VirG, a response regulator which activates ''vir'' promoters. These promoters control genes which are used for infecting the injured plant. | + | The VirA receptor is used by ''A. tumefaciens'' to detect acetosyringone and other phenolic substances which are secreted by plants after injury. In presence of these substances VirA phosphorylates itself and afterwards VirG, a response regulator which activates ''vir'' box containing promoters. These promoters control genes which are used for infecting the injured plant. |
| - | Actually we | + | Actually we tried to use the VirA receptor already existing in the partsregistry (<partinfo>K238008</partinfo>). But due to sequence problems (compare results in [[Team:Bielefeld-Germany/Results/Characterization | characterization]]) we had to isolate the ''virA'' gene from the Ti-plasmid of ''A. tumefaciens'' C58 ourselves and convert it into a BioBrick conform part. We removed an illegal ''PstI'' restriction site in the ''virA'' gene by site-directed mutagenesis. |
You can find this BioBrick here: <partinfo>K389001</partinfo> | You can find this BioBrick here: <partinfo>K389001</partinfo> | ||
| - | You can find | + | You can find the K389001 BioBrick under the control of a constitutive promoter (<partinfo>J23110</partinfo>) here: <partinfo>K389010</partinfo>. |
| - | + | ||
===Mutated ''virG''=== | ===Mutated ''virG''=== | ||
| - | Phosphorylated VirG binds to ''vir'' promoters and activates them. VirG is activated by the acetosyringone receptor VirA. | + | Phosphorylated VirG binds to ''vir'' box containing promoters and activates them. VirG is activated by the acetosyringone receptor VirA. |
| - | This version of VirG activates '' | + | This version of VirG activates ''virB'' promoters in ''Escherichia coli'' without the ''rpoA''-gene from ''Agrobacterium tumefaciens'' [https://2010.igem.org/Team:Bielefeld-Germany/Project/Theory#Subcloning_into_E._coli_and_receptor_function_in_new_host (compare to Theory)]. For this reason the point mutations G56V and I77V are brought into the molecule [http://www.springerlink.com/content/wmq06kua5qkma1au/ YC Jung ''et al.'', (2004)]. Because this BioBrick is synthesized, the codon usage is optimized for ''E. coli'' and illegal restriction sites were removed. When you use this ''virG'' gene in a VirA/G signaling system you do not need <partinfo>K238010</partinfo> anymore to get the system working in ''E. coli''. |
You can find this BioBrick here: <partinfo>K389002</partinfo> | You can find this BioBrick here: <partinfo>K389002</partinfo> | ||
| Line 41: | Line 41: | ||
===''virB''-promoter=== | ===''virB''-promoter=== | ||
''Vir''-promoters from ''A. tumefaciens'' are induced by phosphorylated VirG response regulators and control genes for infecting plants in their natural host. They are part of the VirA/G signal transduction system. | ''Vir''-promoters from ''A. tumefaciens'' are induced by phosphorylated VirG response regulators and control genes for infecting plants in their natural host. They are part of the VirA/G signal transduction system. | ||
| - | We wanted to use the ''vir''-promoter from the partsregistry (<partinfo>K238011</partinfo>) but the same problems occurred | + | We wanted to use the ''vir''-promoter from the partsregistry (<partinfo>K238011</partinfo>) but the same problems occurred as with the use of the VirA receptor BioBrick from the partsregistry. So we also had to create a new ''vir''-promoter BioBrick (again from Ti-plasmid of ''A. tumefaciens'' C58). |
You can find this BioBrick here: <partinfo>K389003</partinfo> | You can find this BioBrick here: <partinfo>K389003</partinfo> | ||
| - | |||
===Firefly luciferase=== | ===Firefly luciferase=== | ||
| - | Bringing the firefly luciferase gene from Promega's pGL4.10[luc2] vector into a BioBrick compatible form as a sensitive reporter gene. To amplify the signal of the luciferase three different sensitivity tuners are assembled before the luciferase gene. The sensitivity tuners were created in 2007 by the iGEM team from Cambridge and amplify the | + | Bringing the firefly luciferase gene from Promega's pGL4.10[luc2] vector into a BioBrick compatible form as a sensitive reporter gene. To amplify the signal of the luciferase three different sensitivity tuners are assembled before the luciferase gene. The sensitivity tuners were created in 2007 by the iGEM team from Cambridge and amplify the readout signal. We also assembled the firefly luciferase behind three different strong constitutive promoters for gathering additional information about this reporter gene [https://2007.igem.org/Cambridge/Amplifier_project#Results Cambridge, (2007)]. |
You can find this BioBrick here: <partinfo>K389004</partinfo> | You can find this BioBrick here: <partinfo>K389004</partinfo> | ||
| Line 70: | Line 69: | ||
===BioBrick for ''virA''-screenings=== | ===BioBrick for ''virA''-screenings=== | ||
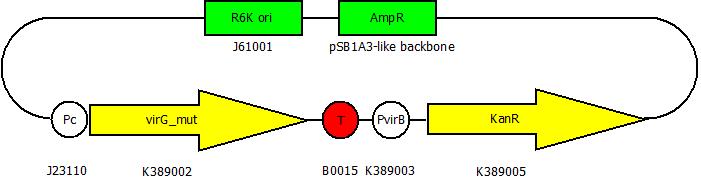
| - | This part contains our mutated ''virG'' BioBrick under the control of a constitutive promoter (<partinfo>J23110</partinfo>) and an antibiotic resistance (<partinfo>K389005</partinfo>) under the control of the ''virB'' promoter (<partinfo>K389003</partinfo>). The better the VirA receptor recognizes a substance the stronger | + | [[Image:Screening_plasmid.jpg|500px|thumb|right|'''Fig. 2: The BioBrick <partinfo>K389011</partinfo> for ''virA'' screenings.''']] |
| + | This part contains our mutated ''virG'' BioBrick under the control of a constitutive promoter (<partinfo>J23110</partinfo>) and an antibiotic resistance (<partinfo>K389005</partinfo>) under the control of the ''virB'' promoter (<partinfo>K389003</partinfo>). The better the VirA receptor recognizes a substance, the stronger the antibiotic resistance will be expressed. | ||
| + | |||
| + | This device is used to screen ''virA'' receptor mutants. The ''virA'' receptor gene is mutated in the <partinfo>BBa_K389010</partinfo> BioBrick. The better the receptor detects a substance, the more VirG is phosphorylated and the stronger the kanamycin resistance from this BioBrick is expressed. The screening system for a mutated ''virA'' gene contains this BioBrick in a plasmid with R6K ori and the BioBrick <partinfo>BBa_K389010</partinfo> in a plasmid with ColE1 ori. Both plasmids are transformed to and screened in ''E. coli'' EC100D because plasmids with R6K ori are only replicated in pir+ or pir116 ''E. coli'' strains. Once a receptor with high sensitivity for a screened substance is found, the plasmids are isolated and transformed to e.g. ''E. coli'' TOP10. Because the R6K ori does not work in this strain because it is not a pir+ / pir116 strain, it is easy to separate the mutated ''virA'' BioBrick from the screening plasmid. | ||
You can find this BioBrick here: <partinfo>K389011</partinfo> | You can find this BioBrick here: <partinfo>K389011</partinfo> | ||
| - | |||
===Reporter constructs=== | ===Reporter constructs=== | ||
| - | The reporter constructs are similar to the ''virA'' screening construct but instead of the antibiotic resistance they carry a reporter gene. The amount of produced reporter shows the activity of the VirA receptor and the ''vir'' promoter, respectively. If the original ''vir'' promoter is too weak, we will use Cambridge's sensitivity tuners to increase the output signal of our biosensor ([ | + | The reporter constructs are similar to the ''virA'' screening construct (compare fig. 2) but instead of the antibiotic resistance they carry a reporter gene. The amount of produced reporter shows the activity of the VirA receptor and the ''vir'' promoter, respectively. If the original ''vir'' promoter is too weak, we will use Cambridge's sensitivity tuners to increase the output signal of our biosensor ([https://2007.igem.org/Cambridge/Amplifier_project#Results Cambridge, 2007]). |
| - | You can find this BioBrick with luciferase | + | You can find this BioBrick with luciferase readout here: <partinfo>K389012</partinfo> |
| - | You can find this BioBrick with mRFP | + | You can find this BioBrick with mRFP readout here: <partinfo>K389013</partinfo> |
| - | You can find this BioBrick with amplified luciferase | + | You can find this BioBrick with amplified luciferase readout No. 1 here: <partinfo>K389411</partinfo> |
| - | You can find this BioBrick with amplified luciferase | + | You can find this BioBrick with amplified luciferase readout No. 2 here: <partinfo>K389412</partinfo> |
| - | You can find this BioBrick with amplified luciferase | + | You can find this BioBrick with amplified luciferase readout No. 3 here: <partinfo>K389413</partinfo> |
===Characterization constructs=== | ===Characterization constructs=== | ||
| - | [[Image:Bielefeld_Vorgehen2.gif|400px|thumb|right|'''Animation of the function of a complete characterization construct with sensitivity tuner (click to enlarge). In the system without sensitivity tuner the reporter gene is directly under the control of the ''vir'' promoter.''' ]] | + | [[Image:Bielefeld_Vorgehen2.gif|400px|thumb|right|'''Fig. 3: Animation of the function of a complete characterization construct with sensitivity tuner (click to enlarge). In the system without sensitivity tuner the reporter gene is directly under the control of the ''vir'' promoter.''' ]] |
| - | The characterization constructs are like the reporter constructs but a ''virA'' gene under the control of a constitutive promoter is assembled before them. These constructs are used to characterize the natural, unmutated VirA/G signaling system and the ''vir'' promoter, respectively. The natural system is induced with acetosyringone and the | + | The characterization constructs are like the reporter constructs but a ''virA'' gene under the control of a constitutive promoter is assembled before them. These constructs are used to characterize the natural, unmutated VirA/G signaling system and the ''vir'' promoter, respectively. The natural system is induced with acetosyringone and the readout is measured to determine the behaviour of the VirA/G signaling system. |
| - | You can find this BioBrick with luciferase | + | You can find this BioBrick with luciferase readout here: <partinfo>K389015</partinfo> |
| - | You can find this BioBrick with mRFP | + | You can find this BioBrick with mRFP readout here: <partinfo>K389016</partinfo> |
| - | You can find this BioBrick with amplified luciferase | + | You can find this BioBrick with amplified luciferase readout No. 1 here: <partinfo>K389421</partinfo> |
| - | You can find this BioBrick with amplified luciferase | + | You can find this BioBrick with amplified luciferase readout No. 2 here: <partinfo>K389422</partinfo> |
| - | You can find this BioBrick with amplified luciferase | + | You can find this BioBrick with amplified luciferase readout No. 3 here: <partinfo>K389423</partinfo> |
===VirA/G signaling system=== | ===VirA/G signaling system=== | ||
| - | A complete VirA/G signaling system without | + | A complete VirA/G signaling system without readout was submitted. The system contains a ''virA'' and our mutated ''virG'' gene under the control of a constitutive promoter and a ''vir'' promoter without reporter gene. |
You can find this BioBrick here: <partinfo>K389017</partinfo> | You can find this BioBrick here: <partinfo>K389017</partinfo> | ||
=References= | =References= | ||
| - | *[ | + | *[https://2007.igem.org/Cambridge/Amplifier_project Cambridge iGEM Team wiki 2007, amplifier project] |
| - | *YC Jung ''et al.'' (2004) | + | *YC Jung ''et al.'', (2004), ''Mutants of ''Agrobacterium tumefaciens virG'' Gene That Activate Transcription of ''vir'' Promoter in ''Escherichia coli'', Current Microbiol 49:334-340. |
Latest revision as of 02:07, 28 October 2010
Contents |
Submitted BioBricks
We have submitted the following working BioBricks. For a more detailed theoretical background to these BioBricks check our theory pages, too.
VirA receptor
The VirA receptor is used by A. tumefaciens to detect acetosyringone and other phenolic substances which are secreted by plants after injury. In presence of these substances VirA phosphorylates itself and afterwards VirG, a response regulator which activates vir box containing promoters. These promoters control genes which are used for infecting the injured plant. Actually we tried to use the VirA receptor already existing in the partsregistry (<partinfo>K238008</partinfo>). But due to sequence problems (compare results in characterization) we had to isolate the virA gene from the Ti-plasmid of A. tumefaciens C58 ourselves and convert it into a BioBrick conform part. We removed an illegal PstI restriction site in the virA gene by site-directed mutagenesis.
You can find this BioBrick here: <partinfo>K389001</partinfo>
You can find the K389001 BioBrick under the control of a constitutive promoter (<partinfo>J23110</partinfo>) here: <partinfo>K389010</partinfo>.
Mutated virG
Phosphorylated VirG binds to vir box containing promoters and activates them. VirG is activated by the acetosyringone receptor VirA. This version of VirG activates virB promoters in Escherichia coli without the rpoA-gene from Agrobacterium tumefaciens (compare to Theory). For this reason the point mutations G56V and I77V are brought into the molecule [http://www.springerlink.com/content/wmq06kua5qkma1au/ YC Jung et al., (2004)]. Because this BioBrick is synthesized, the codon usage is optimized for E. coli and illegal restriction sites were removed. When you use this virG gene in a VirA/G signaling system you do not need <partinfo>K238010</partinfo> anymore to get the system working in E. coli.
You can find this BioBrick here: <partinfo>K389002</partinfo>
virB-promoter
Vir-promoters from A. tumefaciens are induced by phosphorylated VirG response regulators and control genes for infecting plants in their natural host. They are part of the VirA/G signal transduction system. We wanted to use the vir-promoter from the partsregistry (<partinfo>K238011</partinfo>) but the same problems occurred as with the use of the VirA receptor BioBrick from the partsregistry. So we also had to create a new vir-promoter BioBrick (again from Ti-plasmid of A. tumefaciens C58).
You can find this BioBrick here: <partinfo>K389003</partinfo>
Firefly luciferase
Bringing the firefly luciferase gene from Promega's pGL4.10[luc2] vector into a BioBrick compatible form as a sensitive reporter gene. To amplify the signal of the luciferase three different sensitivity tuners are assembled before the luciferase gene. The sensitivity tuners were created in 2007 by the iGEM team from Cambridge and amplify the readout signal. We also assembled the firefly luciferase behind three different strong constitutive promoters for gathering additional information about this reporter gene Cambridge, (2007).
You can find this BioBrick here: <partinfo>K389004</partinfo>
You can find this BioBrick with sensitivity tuner 1 here: <partinfo>K389401</partinfo>
You can find this BioBrick with sensitivity tuner 2 here: <partinfo>K389402</partinfo>
You can find this BioBrick with sensitivity tuner 3 here: <partinfo>K389403</partinfo>
You can find this BioBrick under the control of a weak constitutive promoter here: <partinfo>K389302</partinfo>
You can find this BioBrick under the control of a medium strong constitutive promoter here: <partinfo>K389307</partinfo>
You can find this BioBrick under the control of a strong constitutive promoter here: <partinfo>K389318</partinfo>
Neomycin / kanamycin resistance
A neomycin / kanamycin resistance gene without promoter is isolated and brought into a BioBrick compatible form. We will use the BioBrick <partinfo>P1003</partinfo> as source for the kanamycin resistance gene.
You can find this BioBrick here: <partinfo>K389005</partinfo>
BioBrick for virA-screenings
This part contains our mutated virG BioBrick under the control of a constitutive promoter (<partinfo>J23110</partinfo>) and an antibiotic resistance (<partinfo>K389005</partinfo>) under the control of the virB promoter (<partinfo>K389003</partinfo>). The better the VirA receptor recognizes a substance, the stronger the antibiotic resistance will be expressed.
This device is used to screen virA receptor mutants. The virA receptor gene is mutated in the <partinfo>BBa_K389010</partinfo> BioBrick. The better the receptor detects a substance, the more VirG is phosphorylated and the stronger the kanamycin resistance from this BioBrick is expressed. The screening system for a mutated virA gene contains this BioBrick in a plasmid with R6K ori and the BioBrick <partinfo>BBa_K389010</partinfo> in a plasmid with ColE1 ori. Both plasmids are transformed to and screened in E. coli EC100D because plasmids with R6K ori are only replicated in pir+ or pir116 E. coli strains. Once a receptor with high sensitivity for a screened substance is found, the plasmids are isolated and transformed to e.g. E. coli TOP10. Because the R6K ori does not work in this strain because it is not a pir+ / pir116 strain, it is easy to separate the mutated virA BioBrick from the screening plasmid.
You can find this BioBrick here: <partinfo>K389011</partinfo>
Reporter constructs
The reporter constructs are similar to the virA screening construct (compare fig. 2) but instead of the antibiotic resistance they carry a reporter gene. The amount of produced reporter shows the activity of the VirA receptor and the vir promoter, respectively. If the original vir promoter is too weak, we will use Cambridge's sensitivity tuners to increase the output signal of our biosensor (Cambridge, 2007).
You can find this BioBrick with luciferase readout here: <partinfo>K389012</partinfo>
You can find this BioBrick with mRFP readout here: <partinfo>K389013</partinfo>
You can find this BioBrick with amplified luciferase readout No. 1 here: <partinfo>K389411</partinfo>
You can find this BioBrick with amplified luciferase readout No. 2 here: <partinfo>K389412</partinfo>
You can find this BioBrick with amplified luciferase readout No. 3 here: <partinfo>K389413</partinfo>
Characterization constructs
The characterization constructs are like the reporter constructs but a virA gene under the control of a constitutive promoter is assembled before them. These constructs are used to characterize the natural, unmutated VirA/G signaling system and the vir promoter, respectively. The natural system is induced with acetosyringone and the readout is measured to determine the behaviour of the VirA/G signaling system.
You can find this BioBrick with luciferase readout here: <partinfo>K389015</partinfo>
You can find this BioBrick with mRFP readout here: <partinfo>K389016</partinfo>
You can find this BioBrick with amplified luciferase readout No. 1 here: <partinfo>K389421</partinfo>
You can find this BioBrick with amplified luciferase readout No. 2 here: <partinfo>K389422</partinfo>
You can find this BioBrick with amplified luciferase readout No. 3 here: <partinfo>K389423</partinfo>
VirA/G signaling system
A complete VirA/G signaling system without readout was submitted. The system contains a virA and our mutated virG gene under the control of a constitutive promoter and a vir promoter without reporter gene.
You can find this BioBrick here: <partinfo>K389017</partinfo>
References
- YC Jung et al., (2004), Mutants of Agrobacterium tumefaciens virG Gene That Activate Transcription of vir Promoter in Escherichia coli, Current Microbiol 49:334-340.
 "
"