Team:Bielefeld-Germany/Results/Characterization
From 2010.igem.org
(→K389421, K389422, K389423: Sensitivity Tuner amlified Vir-test system) |
(→K389421, K389422, K389423: Sensitivity Tuner amlified Vir-test system) |
||
| Line 164: | Line 164: | ||
Because there is little difference in induced and not induced system visible and basal transcription rate are high, we assume that the sensitivity tuning constructs are not well applicable for luciferase measurements. | Because there is little difference in induced and not induced system visible and basal transcription rate are high, we assume that the sensitivity tuning constructs are not well applicable for luciferase measurements. | ||
| - | Further Theory click [ | + | Further Theory click [https://2010.igem.org/Team:Bielefeld-Germany/Project/Theory#Read_out_system Read out system] |
=Sequenced BioBricks= | =Sequenced BioBricks= | ||
Revision as of 20:03, 25 October 2010
Tested BioBricks
We tested the following BioBricks:
<partinfo>K238008</partinfo>: virA
We made a restriction analysis and sequenced parts of this BioBrick. There were problems - more information within a short time.
<partinfo>BBa_K238011</partinfo>: vir-promoter
We made a restriction analysis and sequenced parts of this BioBrick.
<partinfo>P1010</partinfo>: ccdB-gene
The ccdB gene targets the gyrase of Escherichia coli and is lethal for all E. coli strains without the gyrase mutation gyrA462 [1]. The ccdB BioBrick is used for the 3A-assembly as a positive selection marker. We transformed this BioBrick into E. coli JM109, DH5α, TOP10, XL1-Blue, EC100D and DB3.1. E. coli JM109, XL1-Blue and DH5α seem to be ccdB resistant because there were as much colonies after P1010 transformation as observed with DB3.1. The P1010 works as expected in E. coli TOP10, EC100D (no colonies after transformation) and DB3.1 (many colonies after transformation).
Table 1: Results of the transformation of the cell-death gene ccdB, BioBrick <partinfo>P1010</partinfo>, into different strains of E. coli.
| E. coli strain | Resistant to ccdB? | Expected result? | Gyrase genotype [2,3] |
|---|---|---|---|
| DB3.1 | yes | yes | gyrA462 |
| DH5α | yes | no | gyrA96 |
| EC100D | no | yes | WT |
| JM109 | yes | no | gyrA96 |
| TOP10 | no | yes | WT |
| XL1-Blue | yes | no | gyrA96 |
It seems that not only the gyrase mutation gyrA462 is causing a ccdB resistance. Also the gyrase mutation gyrA96 gives E. coli a ccdB resistance. This should be kept in mind when assembling BioBricks with the 3A assembly.
<partinfo>K389011</partinfo>: VirA screening device
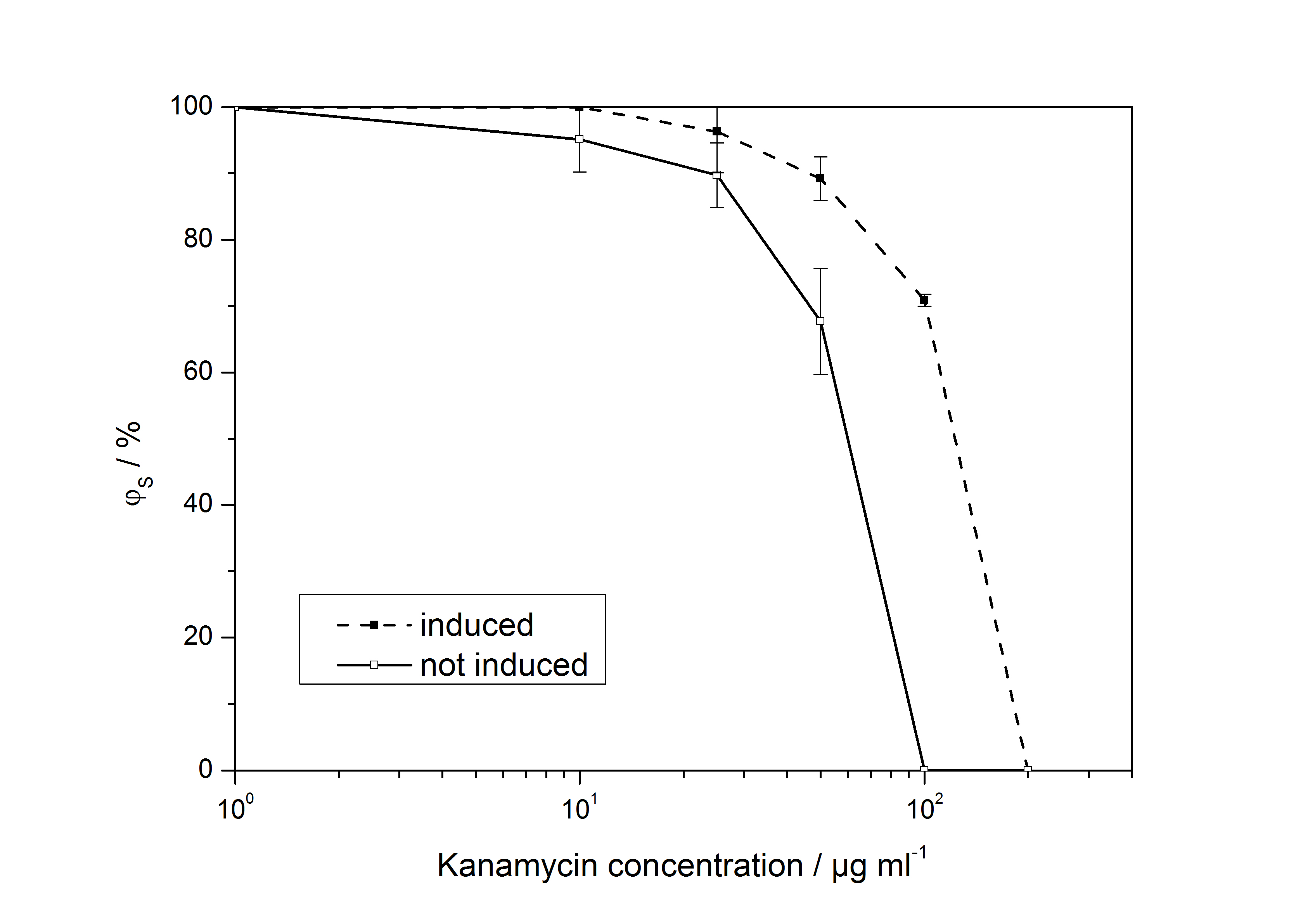
The ratio of surviving colonies ϕS was calculated like
with the number of colony forming units CFU, the concentration of kanamycin on the considered plate KanX and no kanamycin on the plate Kan0.
<partinfo>K389012</partinfo>: VirA reporter system with luciferase
coming more soon
<partinfo>K389015</partinfo>: VirA/G reporter device with luciferase
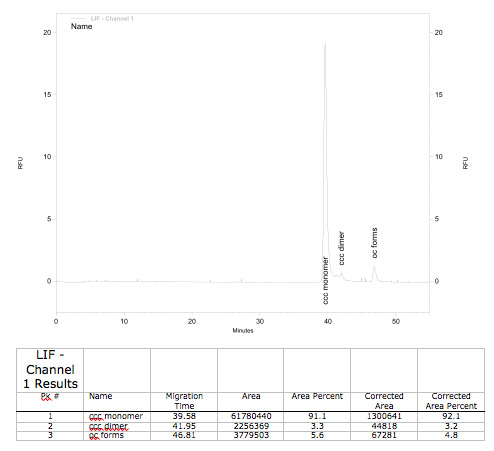
Control of BioBrick quality by capillar gel elektrophoresis (GCE)
<partinfo>K389016</partinfo>: VirA/G reporter device with mRFP
Control of BioBrick quality by capillar gel elektrophoresis (GCE)
<partinfo>K389052</partinfo>: tightly regulated lac operon with mRFP readout
Fails...more soon.
<partinfo>K389421</partinfo>, <partinfo>K389422</partinfo>, <partinfo>K389423</partinfo>: Sensitivity Tuner amlified Vir-test system
By self designed PCR-Primer we excluded terminal GFP and the initial promoter pBAD/araC, for replacing our own VirB promotor and reporter gene luc (luciferase). Primers were designed for sensitivity tuner [http://partsregistry.org/Part:BBa_I746370 II74670], [http://partsregistry.org/Part:BBa_I746380 I746380] and [http://partsregistry.org/Part:BBa_I746390 I746390] so that standard assembly would be possible. Assembling of PCR-products took place by Silver Assembly.
Accomplishment
PCR-Primer Design
Primer forward activator phage P2:
5`-GTT TCT TCG AAT TCG CGG CCG CTT CTA GAT GTT TCA TTG TCC TTT ATG CC-3`
Primer forward activator phage PSP3:
5`-GTT TCT TCG AAT TCG CGG CCG CTT CTA GAT GAT GCA CTG CCC GTT ATG- 3`
Primer forward activator phage phi R73:
5`-GTT TCT TCG AAT TCG CGG CCG CTT CTA GAT GCG CTG CCC TTT CTG-3`
Primer backward Promotor PF from phage P2:
5`-GTT TCT TCC TGC AGC GGC CGC TAC TAG TAT TTC TCC TCT TTC TCT AGT AAG TGG- 3`
Characterization tests
Cultivation was done by induction with Acetosyringone at 50 µM. Controls were not induced Sensitivity Tuner devices as well as induced and not induced nativ system (K389015; without tuning elements). Induction was done upon inoculation. Measuring point for amplification factor calculation was OD 1,0.
Results
Three sensitivity tuned Vir-Gen sensing systems were obtained: K389421, K389422 and K389423 distinguishing by the amplification level of luc transcription.
Space Illustration
Output-signal amplification is in the induced contructs (red) K3894222 and K389423 higher than the not induced controls (green). An exception is K389423 induced with around 200 fold higher amplification than in not induced system. Corresponding to data of iGEM Team, Cambridge 2009, K389423 (originated from I746390) shows the highest amplification rate of all tested Sensitivity Tuners. Our results indicate to a little higher amplification rate of K389421 than K389421 of 100 fold. The controls also show high basal transcription rates.
Because there is little difference in induced and not induced system visible and basal transcription rate are high, we assume that the sensitivity tuning constructs are not well applicable for luciferase measurements.
Further Theory click Read out system
Sequenced BioBricks
Own BioBricks
The sequencing of the following of our own BioBricks was succesful and lead to the expected results:
- <partinfo>K389001</partinfo> (not fully completed)
- <partinfo>K389002</partinfo>
- <partinfo>K389003</partinfo>
- <partinfo>K389004</partinfo>
- <partinfo>K389005</partinfo>
- <partinfo>K389010</partinfo> (not fully completed)
- <partinfo>K389011</partinfo> (not fully completed)
- <partinfo>K389012</partinfo> (not fully completed)
- <partinfo>K389013</partinfo> (not fully completed)
- <partinfo>K389015</partinfo> (not fully completed)
- <partinfo>K389016</partinfo> (not fully completed)
- <partinfo>K389050</partinfo>
Other BioBricks
- <partinfo>K238008</partinfo> sequencing gave negative results - infos in registry are not correct!
- <partinfo>K238011</partinfo> sequencing gave negative results - infos in registry are not correct!
References
[1] http://openwetware.org/wiki/CcdB, CcdB (seen on 10.10.10).
[2] http://openwetware.org/wiki/E._coli_genotypes, E. coli genotypes (seen on 10.10.10).
[3] Metcalf, W.W. et al. (1994) Gene 138, 1.
 "
"