Team:Bielefeld-Germany/Project/Approach
From 2010.igem.org
Contents |
The Approach
First we looked for a sensor system which is able to detect substances and we searched for a substance of interest. The system of interest has to be taken out of a different bacteria species than Escherichia coli, in order to avoid any background. Therefore we checked the literature for a well reviewed sensor system, which is not part of the E. coli genome. The system of choice was the phenolic sensing system virA of Agrobacterium tumefaciens. It naturally detects acetosyringone, which is a secondary metabolite of plants that affects bacteria as an attractant. After repeated research to check for any other possible substance which could be detected by the system, we got a really long list of possibilities and picked 'capsaicin', which is responsible for the spiciness of edibles.
The biological steps for creating a new sensor system are:
- extract the virA system out of A. tumefaciens
- create new BioBricks out of the exciting environmental parts
- transform the new BioBricks into E. coli
- modify the system for sensibility and specificity by error prone PCR
- find and select the most promising mutants
Preparing the system
We tried to work with the already existing virA from the registry (<partinfo>K238008</partinfo>). Unfortunately this virA BioBrick did not work. The sequence data of the BioBrick did not fit to the published virA sequence. So we had to extract the virA gene via PCR out of the A. tumefaciens TI-plasmid by ourselves in order to create a new BioBrick.
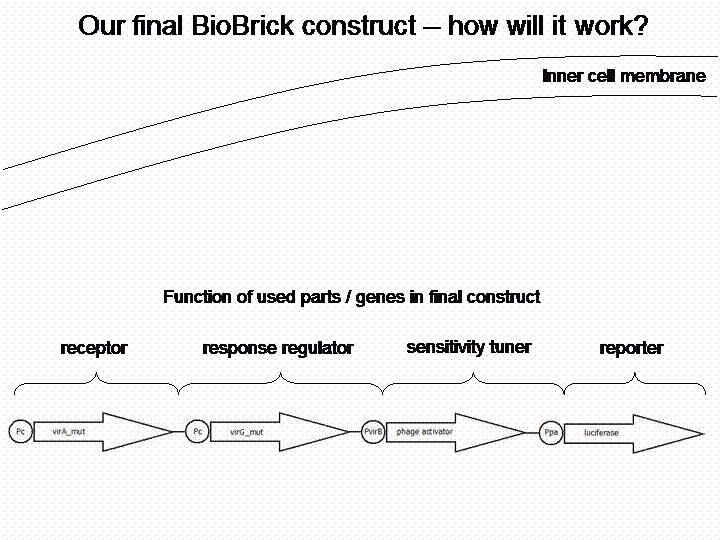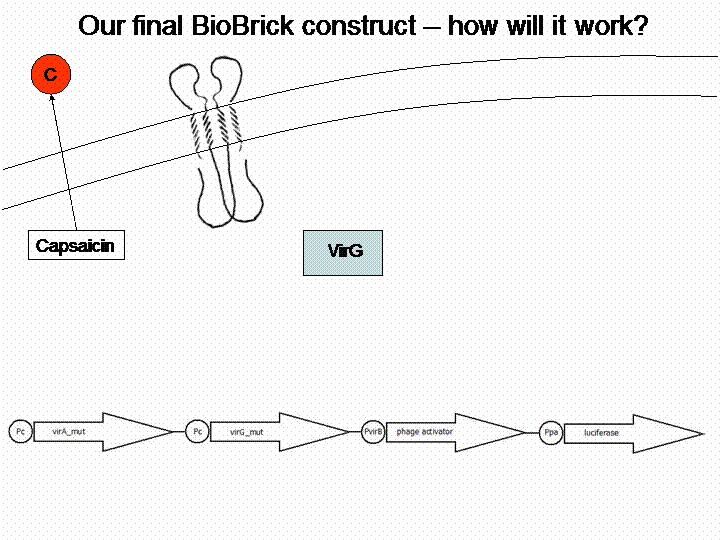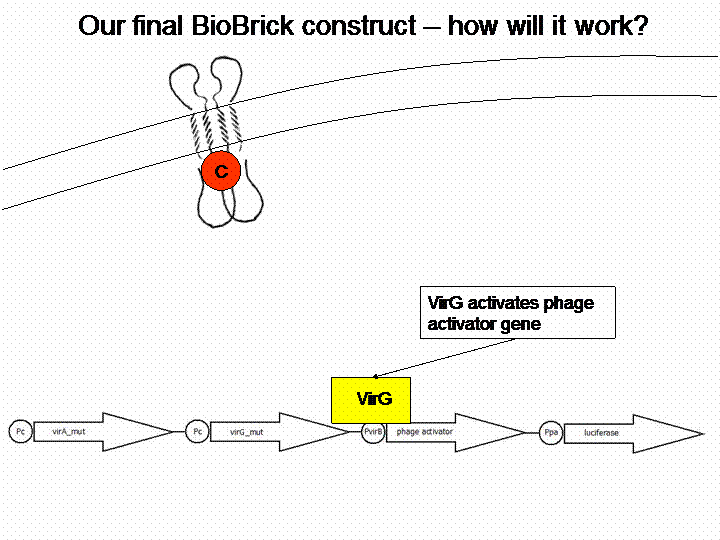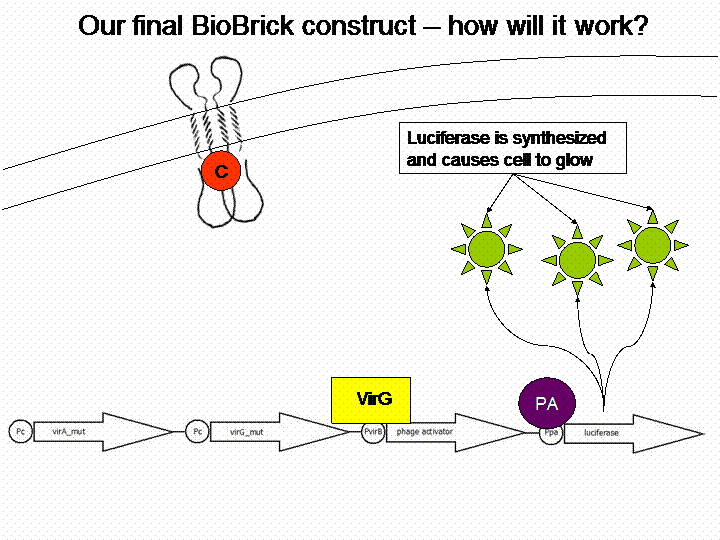
VirA does not work without the help of the VirG protein. The existing virG gene in the iGEM registry contains illegal restriction sites. Moreover it needs the help of ChvE Protein and sugars to work perfectly with VirA. We changed the sequence manually and had the gene synthesized by Mr. Gene.
Starting point for biobricks -> Go cloning
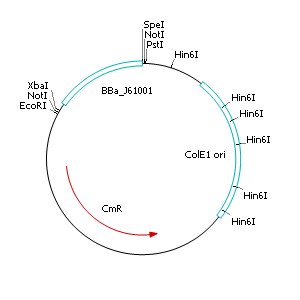
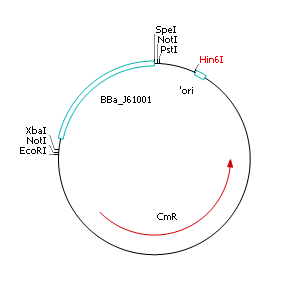
The applied screening system in E. coli consists of two plasmids. After discussing the possibility of creating only one big plasmid, we unifed that one plasmid would minimize transformation efficiency and would be difficult to modify via error prone PCR. So we had to change the origin of a pSB1X3 plasmid in order to avoid incompatibility. Thus we cloned the R6K origin (<partinfo>J61001</partinfo>) into the <partinfo>pSB1C3</partinfo> plasmid and subsequently removed the original pMB1/ColE1 origin of replication. Therefore we digested the pSB1C3::R6K plasmid with Hin6I and seperated the resulting 6 fragments (1758 bp, 270 bp, 174 bp, 109 bp, 100 bp, 67 bp) by agarose gel electrophoresis. We extracted the largest fragment (1758 bp) and religated it. The resulting plasmid still contains 32 remaining basepairs of the pMB1 origin, but this fragment does not enable replication in pir- strains.
The error prone PCR
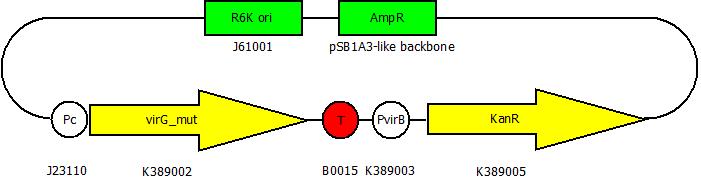
After cloning the new origin into the pSBXXX-backbones we were able to create the two constructs. The first one will be found inside the competent bacteria cells and contains the virG gene under the constitutive promotor <partinfo>J23110</partinfo>, a terminator (<partinfo>B0017</partinfo>), a vir promotor and a readout or selection gene (luciferase, mRFP and kanamycin resistance, respectively):
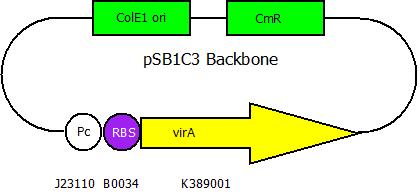
The second plasmid contains the virA gene under the control of the constitutive promotor <partinfo>J23110</partinfo> and will be transformed and modified in one step via error prone PCR.
The error prone PCR is a PCR under malfunction conditions for the polymerase. It is possible to regulate the frequency of the mutagenesis by editing unbalanced concentrations of nucleotides and co-factors to the PCR. So we are going to create a new BioBrick and transform it into our target in one step. Before we are able to do the error prone PCR we need to get the backbones done.
Directed mutagenesis
VirA_mut1 Regarding to the results from Muscle, we used site directed mutagenises (for Protocols see: ([http://openwetware.org/wiki/Site-directed_mutagenesis OpenWet Ware])) to change VirA protein sequence leucin 293 to tyrosine (L293Y = mut1).
VirA_mut2
After VirA_mut1 with its single mutation failed working in our screening system, we decided to mutagenize some further amino acids in the area of the reported binding region from the animal TRPV1 receptors.
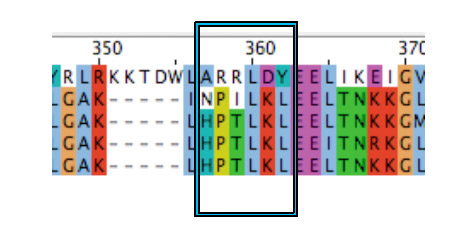
According to the literature amino acid residues from 288 to 293, regulate phenol structural specificity in the TRPV1 receptor. Substitutions in this region narrow the active phenols to those missing the para-acyl substituent ([http://www.springerlink.com/content/978-0-387-78884-5#section=169995&page=1 Lin et al., 2008]). Our VirA_mut2 construct contains the following substitutions:
- A357H
- R358P
- R359T
- L360L
- D361K
- Y361L
Survival of the fittest
We use high amounts of the antibiotic kanamycin in order to select the most specific system for our substances. By varying the amount of kanamycin we will be able to carefully select the best mutant out of our PCR-tests. The mutated virA system will be induced after the error prone PCR by a mix of possible targets for the system (like capcaicin, dopamin, homovanillic acid etc.). It is important to avoid any acetosyringone, so we will be able to search for systems with new targets.
The final construct
Sensitivity Tuner amlified Vir-test system: <partinfo>K389421</partinfo>, <partinfo>K389422</partinfo>, <partinfo>K389423</partinfo>:
By self designed PCR-Primer we excluded terminal GFP and the initial promoter pBAD/araC, for replacing our own VirB promotor and reporter gene luc (luciferase). Primers were designed for sensitivity tuner [http://partsregistry.org/Part:BBa_I746370 I746370], [http://partsregistry.org/Part:BBa_I746380 I746380] and [http://partsregistry.org/Part:BBa_I746390 I746390] so that standard assembly would be possible. The RBS from the original Sensitivity Tuner for the phage activator war thereby removed. New RBS comes with Virb-promotor element [http://partsregistry.org/cgi/sequencing/one_blast.cgi?id=7888 K389003]. Assembling of PCR-products took place by Silver Assembly.
Accomplishment
PCR-Primer Design
Primer forward activator phage P2:
5`-GTT TCT TCG AAT TCG CGG CCG CTT CTA GAT GTT TCA TTG TCC TTT ATG CC-3`
Primer forward activator phage PSP3:
5`-GTT TCT TCG AAT TCG CGG CCG CTT CTA GAT GAT GCA CTG CCC GTT ATG- 3`
Primer forward activator phage phi R73:
5`-GTT TCT TCG AAT TCG CGG CCG CTT CTA GAT GCG CTG CCC TTT CTG-3`
Primer backward Promotor PF from phage P2:
5`-GTT TCT TCC TGC AGC GGC CGC TAC TAG TAT TTC TCC TCT TTC TCT AGT AAG TGG- 3`
Characterization tests
Cultivation was done by induction with Acetosyringone at 50 µM. Controls were not induced Sensitivity Tuner devices as well as induced and not induced nativ system ([http://partsregistry.org/Part:BBa_K389015 K389015]; without tuning elements). Induction was done upon inoculation. Measuring point for amplification factor calculation was OD 1.0.
For demonstration of our final system see the animation below:
 "
"