Team:LMU-Munich/Notebook/Apoptosis
From 2010.igem.org
(→8-17-2010) |
(→What we did) |
||
| (484 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
{{:Team:LMU-Munich/Templates/Page Header}} | {{:Team:LMU-Munich/Templates/Page Header}} | ||
| - | = | + | ==<font color="#9933CC">'''ApoControl Notebook'''</font>== |
| - | ==''' | + | |
| - | {| class="wikitable centered" border="2" rules="rows" width="100%" style="border-color:white;" | + | == '''What we did''' == |
| + | <b>Short description of our work, our results and our supporters</b> | ||
| + | |||
| + | |||
| + | The creation of certain constructs was necessary for our two systems for cell selection by means of apoptosis: “Cut’N’Survive” and “Jump-Or-Die”. We searched for sources of the DNA sequences we needed and found several supporters which are listed below. | ||
| + | |||
| + | Most genes and promoters were amplificated via PCR with overhang-primers with the BioBrick prefix or suffix. If the sequence contained a EcoR1-, Pst1-, Xba1-, Spe1- or Not1- restriction site, we used mutagenesis primers and fusioned both DNA parts by fusion PCR. All PCRs worked out, even the fusion PCRs. | ||
| + | |||
| + | The length of the PCR products were tested by agarose gel electrophoresis. We tried to sequence our PCR products, but obtained poor results and resorted to sequencing the plasmids. | ||
| + | |||
| + | In parallel, we made competent cells and multiplied ccdB (death gene)-vectors with different antibiotic resistances. All components were digested with the appropriate restriction enzymes. The samples were cleaned with a PCR clean up kit or dephosphorylated to reduce false ligations. | ||
| + | |||
| + | We ligated our constructs and several interim stages with the 3A-assembly according to our schedule. The ligations were transformed to E.coli DH5α strains and selected by antibiotics. Afterwards, some colonies were picked and we tested the insertion of the construct by colony PCR. | ||
| + | |||
| + | If the colony PCR resulted in bands of the right size, we extracted the plasmids from overnight cultures and sequenced the samples with forward and reverse BioBrick primers. | ||
| + | |||
| + | Unfortunately, not all BioBricks were cloned succesfully. However, we were able to produce 4 BioBricks, one of which represents a full construct while the other three are intermediates. The system wasn't completed on time, so we weren´t able to test them in eukarytic cell lines. | ||
| + | |||
| + | |||
| + | |||
| + | <b>The protocols we used are listed here: </b> [[Team:LMU-Munich/Notebook#Protocols|Protocols]] | ||
| + | |||
| + | <b>These Biobricks we submitted: </b> | ||
| + | |||
| + | *BBa_K368004: attP+eGFP+SV40PA | ||
| + | *BBa_K368011: eGFP+SV40PA | ||
| + | *BBa_K368016: TEVrecognition site+N-degron+SF3b155 | ||
| + | *BBa_K368019: TEV-Protease+p14*+TEVrecognition site | ||
| + | |||
| + | <b>Sources, helpers and supporters:</b> | ||
| + | |||
| + | * Prof. Dr. Angelika Böttger : | ||
| + | ** prevTRE (tet-on CMV promoter; inducible by doxycycline in special cell lines) | ||
| + | ** supported the construction ideas and would have given us the cells and mediums we would have needed | ||
| + | ** SV40PA (Polyadenylation site): gave us a vector containing it | ||
| + | ** Human Bak: her assistant Erika Clement gave us appropriate cDNA | ||
| + | |||
| + | * Dr. Arnim Weber: submitted us a vector with human Bak | ||
| + | * Dr. Philipe Soriano: <html> | ||
| + | <a href="http://www.ncbi.nlm.nih.gov/pubmed/17225864?dopt=Abstract"> (Raymond CS et al: High-Efficiency FLP and PhiC31 Site-Specific Recombination in Mammalian Cells (2007))</a> | ||
| + | </html> | ||
| + | ** Sequences of attB and attP site | ||
| + | ** PhiC31o was bought via addgene | ||
| + | * Knop, M (Heidelberg): <html> | ||
| + | <a href ="http://www.ncbi.nlm.nih.gov/pubmed?term=Efficient%20protein%20depletion%20by%20genetically%20controlled%20deprotection%20of%20a%20dormant%20N-degron">(Knop et al.: Efficient protein depletion by genetically controlled deprotection of a dormant N-degron (2009))</a> | ||
| + | </html> | ||
| + | ** TEVrecognition site+N-degron+SF3b155 | ||
| + | ** TEV-Protease+p14*+TEVrecognition site | ||
| + | * Prof. Dr. Thorsten Mascher: | ||
| + | ** Helped with primer design, agarose gel electrophoresis apparatuses and trouble shooting | ||
| + | * Prof. Dr. Kirsten Jung: | ||
| + | ** Helped with ideas and fundraising | ||
| + | * Dr. Susanne Gebhard: | ||
| + | ** Helped with trouble-shooting and materials | ||
| + | * Prof. Dr. Andreas Brachmann: | ||
| + | ** Sequenced our samples | ||
| + | * Partsregistry: | ||
| + | ** eGFP (BBa_I714891) | ||
| + | ** CMV-Promoter (BBa_J52034: this part was wrong: its lacI !!!) | ||
| + | ** ccdB amp, cam, tet, kan in E.coli DH3 | ||
| + | |||
| + | =='''Contents'''>== | ||
| + | |||
| + | |||
| + | {|class="wikitable centered" border="2" rules="rows" width="100%" style="border-color:white;" | ||
|- | |- | ||
! Week !! colspan="7" |Days | ! Week !! colspan="7" |Days | ||
| + | |- | ||
| + | | | ||
| + | |style="text-align:center"|Monday | ||
| + | |style="text-align:center"|Tuesday | ||
| + | |style="text-align:center"|Wednesday | ||
| + | |style="text-align:center"|Thursday | ||
| + | |style="text-align:center"|Friday | ||
| + | |style="text-align:center"|Saturday | ||
| + | |style="text-align:center"|Sunday | ||
|- | |- | ||
|style="text-align:center"| 31 | |style="text-align:center"| 31 | ||
| Line 88: | Line 160: | ||
|style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-02-2010|10-02-2010]] | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-02-2010|10-02-2010]] | ||
|style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-03-2010|10-03-2010]] | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-03-2010|10-03-2010]] | ||
| + | |- | ||
| + | |style="text-align:center"| 40 | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-04-2010|10-04-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-05-2010|10-05-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-05-2010|10-06-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-07-2010|10-07-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-08-2010|10-08-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-09-2010|10-09-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-10-2010|10-10-2010]] | ||
| + | |- | ||
| + | |style="text-align:center"| 41 | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-11-2010|10-11-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-12-2010|10-12-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-13-2010|10-13-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-14-2010|10-14-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-15-2010|10-15-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-16-2010|10-16-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-17-2010|10-17-2010]] | ||
| + | |- | ||
| + | |style="text-align:center"| 42 | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-18-2010|10-18-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-19-2010|10-19-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-20-2010|10-20-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-21-2010|10-21-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-22-2010|10-22-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-23-2010|10-23-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-24-2010|10-24-2010]] | ||
| + | |- | ||
| + | |style="text-align:center"| 43 | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-25-2010|10-25-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-26-2010|10-26-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-27-2010|10-27-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-28-2010|10-28-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-29-2010|10-29-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-30-2010|10-30-2010]] | ||
| + | |style="text-align:center"| [[Team:LMU-Munich/Notebook/Apoptosis#10-31-2010|10-31-2010]] | ||
|- | |- | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
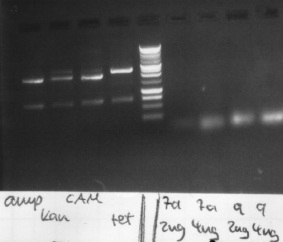
== 8-10-2010 == | == 8-10-2010 == | ||
| Line 205: | Line 240: | ||
- 3 ml LB-Media + 4 µl Kanamycin | - 3 ml LB-Media + 4 µl Kanamycin | ||
| - | - Inoculated | + | - Inoculated with 1 colony of BBa_I714891 -> 37°C |
| Line 215: | Line 250: | ||
| - | <font color="#009933">Restriction | + | <font color="#009933">Restriction digestion of CMV-Promoter BBa_J52034 with EcoRI and PstI</font> |
{| | {| | ||
| Line 259: | Line 294: | ||
- 10µg DNA: pDS7 (2µl), eGFP (15µl), 190-6 (10µl) | - 10µg DNA: pDS7 (2µl), eGFP (15µl), 190-6 (10µl) | ||
| - | <font color="#009933">Plate colonies for plasmid | + | <font color="#009933">Plate colonies for plasmid extraction</font> |
- CMV (Kanamycin), eGFP (Kanamycin), pDS7 (Ampicillin), 190-6 (Ampicillin)) | - CMV (Kanamycin), eGFP (Kanamycin), pDS7 (Ampicillin), 190-6 (Ampicillin)) | ||
| Line 273: | Line 308: | ||
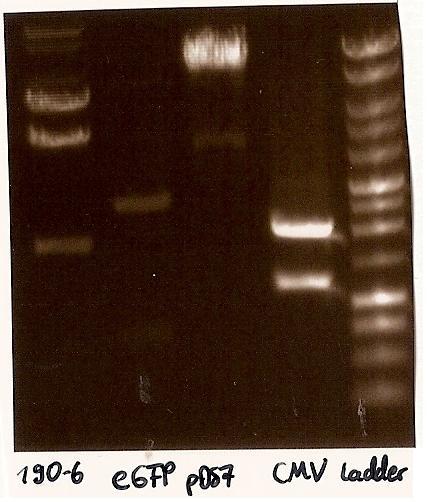
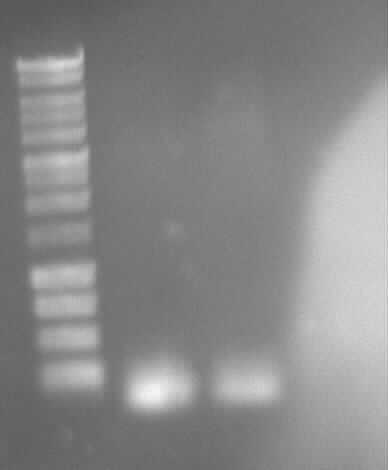
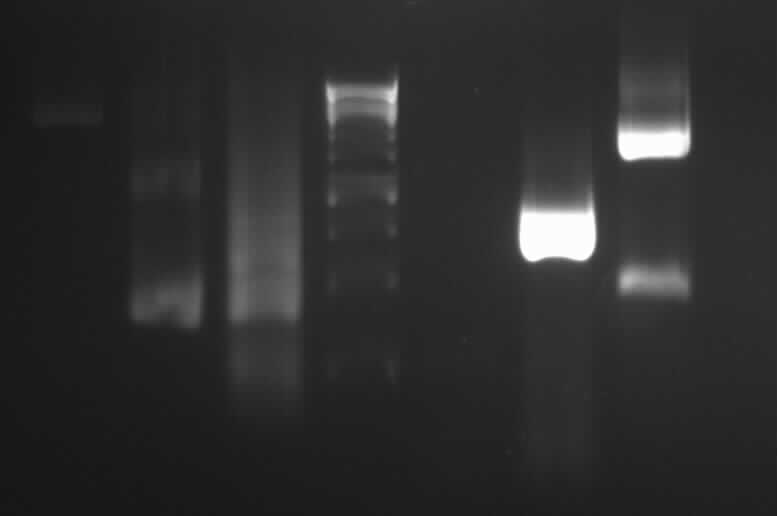
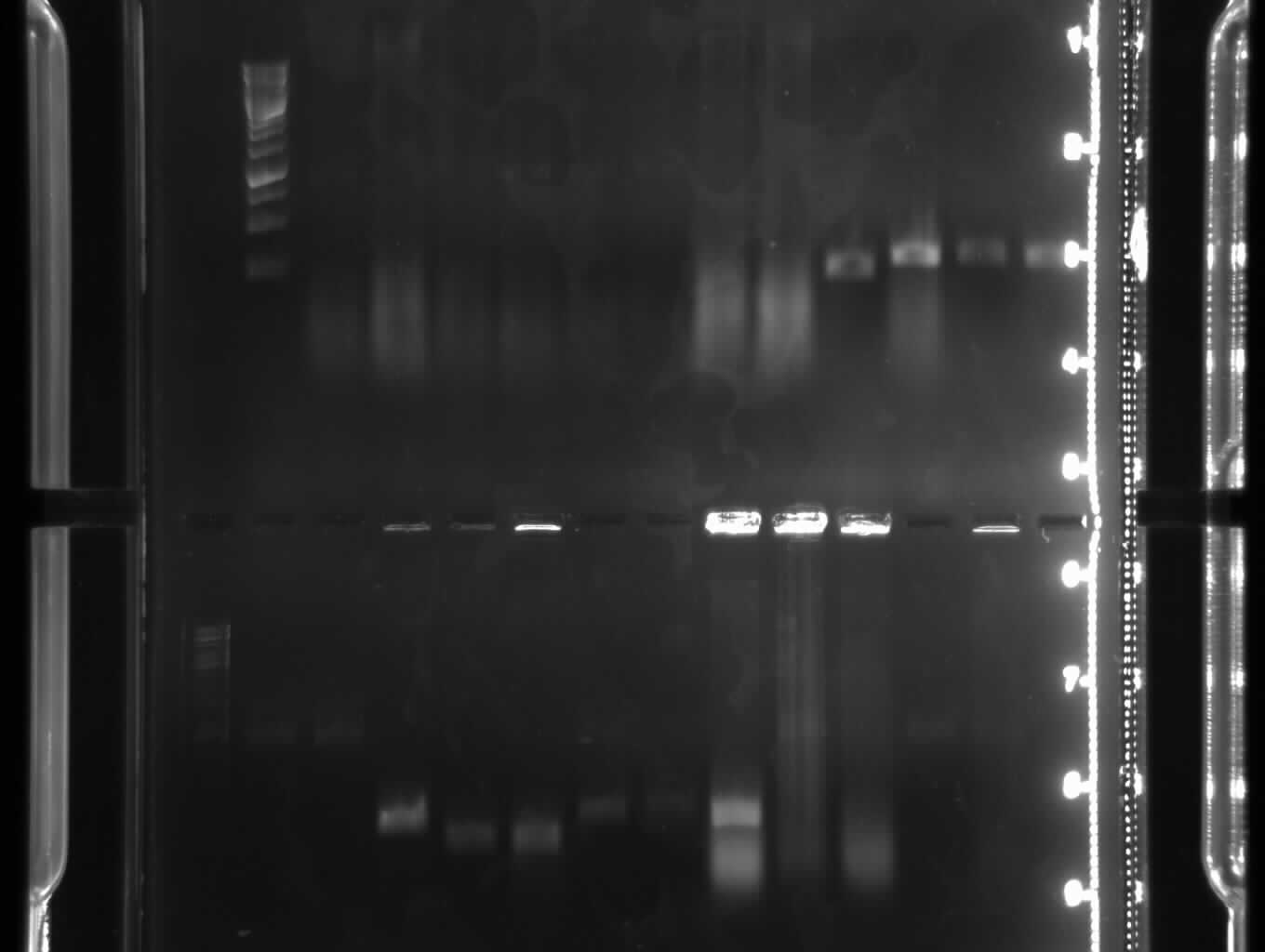
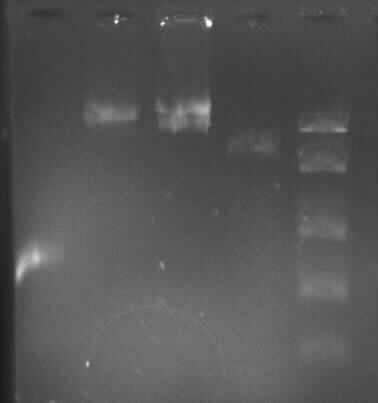
[[Image:GelverdauCMVpDS71906eGFP.jpg|thumb|right|Gelfoto from the EcoR1 and Pst1 Restrictiondigest of 190-6, eGFP, pDS7 and CMV]] | [[Image:GelverdauCMVpDS71906eGFP.jpg|thumb|right|Gelfoto from the EcoR1 and Pst1 Restrictiondigest of 190-6, eGFP, pDS7 and CMV]] | ||
| - | <font color="#009933">Inoculate CMV into LB medium with | + | <font color="#009933">Inoculate CMV into LB medium with ampicillin</font> |
- CMV (BBa_J52034) from 10.8.2010 inoculated into LB medium with ampicillin, as falsly inoculated in Kanamycin | - CMV (BBa_J52034) from 10.8.2010 inoculated into LB medium with ampicillin, as falsly inoculated in Kanamycin | ||
| Line 285: | Line 320: | ||
- expected DNA bands: 190-6 (4840bp, 1903bp), pDS7 (8027bp, 6bp), CMV (654 bp (Insert), 2079bp (Plasmid)), eGFP (720bp (Insert), 2750bp (Plasmid)) | - expected DNA bands: 190-6 (4840bp, 1903bp), pDS7 (8027bp, 6bp), CMV (654 bp (Insert), 2079bp (Plasmid)), eGFP (720bp (Insert), 2750bp (Plasmid)) | ||
| - | - Correct DNA bands for 190-6 (~4800bp, ~1900bp, ~6700bp (undigested plasmid)) and eGFP (~2000bp (Plasmid), ~750 bp (Insert)); CMV probably not digested (two bands; one probably normal, one supercoiled) and pDS7 not clear | + | - <font color="#CC33CC">'''Correct DNA bands for 190-6 (~4800bp, ~1900bp, ~6700bp (undigested plasmid)) and eGFP (~2000bp (Plasmid), ~750 bp (Insert))'''</font>; CMV probably not digested (two bands; one probably normal, one supercoiled) and pDS7 not clear |
<font color="#009933">Restriction digest from CMV and pDS7</font> | <font color="#009933">Restriction digest from CMV and pDS7</font> | ||
| Line 301: | Line 336: | ||
- Expected DNA bands: CMV see above, pDS7 (3647bp, 3369bp, 1011bp, 6bp) | - Expected DNA bands: CMV see above, pDS7 (3647bp, 3369bp, 1011bp, 6bp) | ||
| - | - false DNA bands CMV (~1200 bp, ~2000 bp) and pDS7 (~8000bp two bands, ~1100 bp); required to isolate a new colony for these two Plasmidextractions | + | - false DNA bands CMV (~1200 bp, ~2000 bp) and pDS7 (~8000bp two bands, ~1100 bp); required to isolate a new colony for these two Plasmidextractions |
<font color="#009933">Plated CMV on Ampicllin-Agar</font> | <font color="#009933">Plated CMV on Ampicllin-Agar</font> | ||
| Line 377: | Line 412: | ||
|- | |- | ||
| | | | ||
| - | |||
|pTRERev (0,15µg/µl) | |pTRERev (0,15µg/µl) | ||
| + | |pcDNA3 (0,6 µg/µl) | ||
|- | |- | ||
| Line 485: | Line 520: | ||
<font color="#009933">Agarose gel electrophoresis</font> | <font color="#009933">Agarose gel electrophoresis</font> | ||
| - | ->Protocol ([[Team:LMU-Munich/Notebook/Protocols/11_Agarose_gel_electrophoresis|11 Agarose gel electrophoresis]]) | + | -> Protocol ([[Team:LMU-Munich/Notebook/Protocols/11_Agarose_gel_electrophoresis|11 Agarose gel electrophoresis]]) |
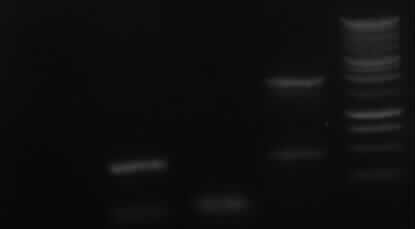
- Agarose gel electrophoresis of the restriction digest of PhiC31o and PCR 1 and 6 | - Agarose gel electrophoresis of the restriction digest of PhiC31o and PCR 1 and 6 | ||
| - | - the right bands found for PhiC31o (~2900,~2400,~250) | + | - <font color="#CC33CC">'''the right bands found for PhiC31o (~2900,~2400,~250)'''</font> |
| - | - the right band found for PCR1 (~450) | + | - <font color="#CC33CC">'''the right band found for PCR1 (~450)'''</font> |
- no band found for PCR6; new electrophoresis needed with more DNA loaded | - no band found for PCR6; new electrophoresis needed with more DNA loaded | ||
| Line 518: | Line 553: | ||
- new agarose gel electrophoresis from PCR6 with 5µl DNA instead of 3µl (image not yet shown) | - new agarose gel electrophoresis from PCR6 with 5µl DNA instead of 3µl (image not yet shown) | ||
| - | - the right band found for PCR6 (~200) | + | -<font color="#CC33CC"> '''the right band found for PCR6 (~200)'''</font> |
<font color="#009933">New overnight cultures of CMV and pDS7</font> | <font color="#009933">New overnight cultures of CMV and pDS7</font> | ||
| Line 525: | Line 560: | ||
<font color="#009933">PCR purification of PCR 1 and 6</font> | <font color="#009933">PCR purification of PCR 1 and 6</font> | ||
| + | |||
| + | -> Protocol ([[Team:LMU-Munich/Notebook/Protocols/12_Gel_extraction_or_PCR_Clean_up|12 Gel extraction or PCR Clean up]]) | ||
- DNA concentration of the PCR 1 and 6 products measured: PCR1: 410ng/µl (A260/A280=1.253) PCR6: 568ng/µl (A260/A280=1.275) | - DNA concentration of the PCR 1 and 6 products measured: PCR1: 410ng/µl (A260/A280=1.253) PCR6: 568ng/µl (A260/A280=1.275) | ||
| Line 533: | Line 570: | ||
-> PCR6: 37.5ng/µl (A260/A280=1.667) | -> PCR6: 37.5ng/µl (A260/A280=1.667) | ||
| - | |||
| - | |||
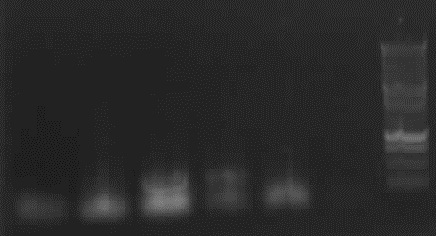
== 8-18-2010 == | == 8-18-2010 == | ||
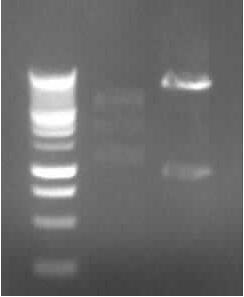
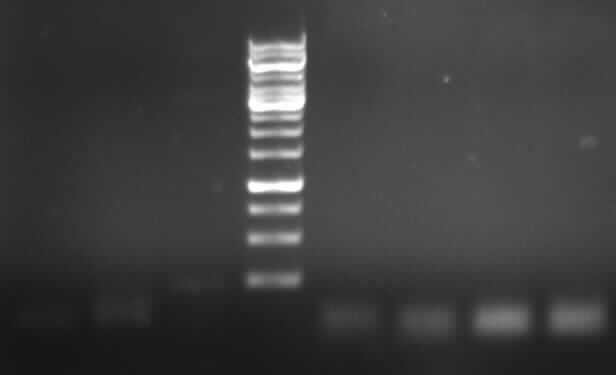
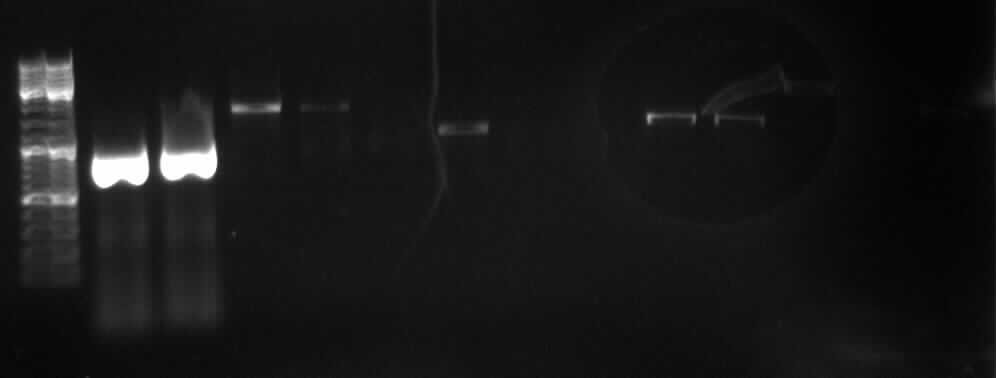
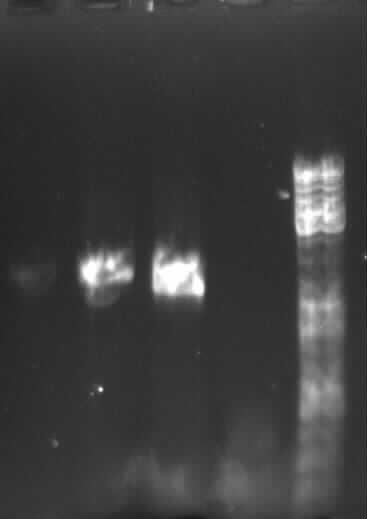
[[Image:18_8_10_apo.jpg|thumb|right|Agarose gel electrophoresis of (from left to right) CMV and pDS7 showing the right bands for pDS7]] | [[Image:18_8_10_apo.jpg|thumb|right|Agarose gel electrophoresis of (from left to right) CMV and pDS7 showing the right bands for pDS7]] | ||
| - | + | <font color="#009933">Plasmid Extraction of CMV and pDS7</font> | |
-> Protocol ([[Team:LMU-Munich/Notebook/Protocols/4_Plasmid_extraction_from_cells|4 Plasmid extraction from cells]]) | -> Protocol ([[Team:LMU-Munich/Notebook/Protocols/4_Plasmid_extraction_from_cells|4 Plasmid extraction from cells]]) | ||
| - | - | + | - Plasmid extraction of CMV (97.5ng/µl; A260/A280=1.857) and pDS7 (212ng/µl; A260/A280=1.848) |
| - | + | <font color="#009933">Restriction digestion</font> | |
| - | -> Protocol ([[Team:LMU-Munich/Notebook/Protocols/5_Restriction_digest|5 Restriction | + | -> Protocol ([[Team:LMU-Munich/Notebook/Protocols/5_Restriction_digest|5 Restriction digestion]]) |
| - | - Agarose Gel electrophoresis of digested CMV and pDS7 | + | - Restriction digestion of CMV (EcoR1 + Pst1; 10µl DNA, buffer H) and pDS7 (EcoR1 + Spe1; 5µl DNA, buffer B) |
| + | |||
| + | -> expected DNA bands: CMV: 2079bp (plasmid) + 654bp (Insert); pDS7: 7022bp + 1011bp | ||
| + | |||
| + | <font color="#009933">Agarose Gel electrophoresis of digested CMV and pDS7</font> | ||
-> Protocol ([[Team:LMU-Munich/Notebook/Protocols/11_Agarose_gel_electrophoresis|11 Agarorse gel electrophoresis]]) | -> Protocol ([[Team:LMU-Munich/Notebook/Protocols/11_Agarose_gel_electrophoresis|11 Agarorse gel electrophoresis]]) | ||
| - | -> right DNA bands for pDS7 (~7000bp, ~1000bp) | + | -> <font color="#CC33CC">'''right DNA bands for pDS7 (~7000bp, ~1000bp)'''</font> |
-> false DNA bands for CMV | -> false DNA bands for CMV | ||
| - | - Starting PCR 2a and 2b (replication and mutagenesis of pDS7): 3 µl DNA and 50°C Annealing Temperatur (other same as | + | - Starting PCR 2a and 2b (replication and mutagenesis of pDS7): 3 µl DNA and 50°C Annealing Temperatur (other same as 8-16-2010) |
== 8-19-2010 == | == 8-19-2010 == | ||
| - | + | ||
| + | <font color="#009933">Agarose gel electrophoresis of PCR 2a and 2b</font> | ||
| + | |||
| + | -> Protocol ([[Team:LMU-Munich/Notebook/Protocols/11_Agarose_gel_electrophoresis|11 Agarose gel electrophoresis]]) | ||
| + | (150V, 30min) | ||
{| | {| | ||
| Line 568: | Line 611: | ||
|} | |} | ||
| - | -> the right bands for PCR2a (~300bp) and PCR2b (~700bp) | + | -><font color="#CC33CC"> '''the right bands for PCR2a (~300bp) and PCR2b (~700bp)'''</font> |
- New agarose gel electrophoresis with all of the PCR product for gel extraction (150V, 30min) | - New agarose gel electrophoresis with all of the PCR product for gel extraction (150V, 30min) | ||
| - | + | <font color="#009933">Gel extraction of the DNA from PCR2a and PCR2b</font> | |
| - | + | ||
| - | + | ||
-> Protocol ([[Team:LMU-Munich/Notebook/Protocols/12_Gel_extraction_or_PCR_Clean_up|12 Gel extraction or PCR Clean up]]) | -> Protocol ([[Team:LMU-Munich/Notebook/Protocols/12_Gel_extraction_or_PCR_Clean_up|12 Gel extraction or PCR Clean up]]) | ||
| Line 580: | Line 621: | ||
- DNA concentration measured; problem with nanodrop as too low concentration; lyophille used to reduce volume | - DNA concentration measured; problem with nanodrop as too low concentration; lyophille used to reduce volume | ||
| - | - DNA concentration measured again: PCR2a: 70ng/µl A260/A280=1.647; PCR2b: 45ng/µl A260/A280=1.5 | + | - DNA concentration measured again: PCR2a: 70ng/µl A260/A280=1.647; PCR2b: 45ng/µl A260/A280=1.5 |
| + | |||
| + | <font color="#009933">PCR 3 (joining PCR of 2a and 2b)</font> | ||
- PCR3 (the joining PCR of PCR2a and 2b; Joining of the TEVrecogn-N-Degron-SF3 part) done: 1.3 µl of PCR2a and 4.7 µl of PCR2b makes 300ng of a 1:1 solution of both to be joined DNA parts. Annealing temperature: 50°C | - PCR3 (the joining PCR of PCR2a and 2b; Joining of the TEVrecogn-N-Degron-SF3 part) done: 1.3 µl of PCR2a and 4.7 µl of PCR2b makes 300ng of a 1:1 solution of both to be joined DNA parts. Annealing temperature: 50°C | ||
| Line 587: | Line 630: | ||
== 8-20-2010 == | == 8-20-2010 == | ||
| - | + | <font color="#009933">Agarose gel electrophoresis of PCR3</font> | |
| + | |||
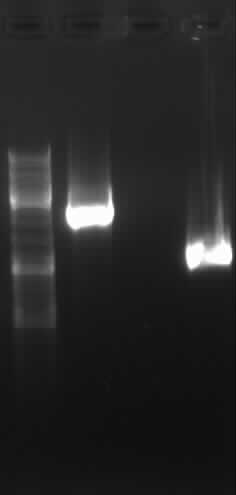
| + | [[Image:GelverdauPCR3-1.jpg|400px|Gelfoto from PCR3]] | ||
| + | |||
| + | left column: marker; rightmost column: PCR3 | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] (150V, 30min) | ||
| + | |||
| + | -> expected band: ~1000bp | ||
| + | |||
| + | -> false band: ~500bp | ||
| + | |||
| + | - probable reason: mini photometre was influenced by gel extraction chemicals, therefore it measured false DNA concentrations and false template masses were calculated | ||
| + | |||
| + | -> New 2a and 2b PCR | ||
| + | |||
| + | <font color="#009933">New PCR (2a and 2b)</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | (see 8-18-2010, but 35,5µl water) | ||
== 8-21-2010 == | == 8-21-2010 == | ||
| - | + | weekend | |
== 8-22-2010 == | == 8-22-2010 == | ||
| - | + | weekend | |
== 8-23-2010 == | == 8-23-2010 == | ||
| - | + | ||
| + | <font color="#009933">Agarose gel electrophoresis of PCR 2b</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | - expected band: 700bp | ||
| + | |||
| + | -> no band shown on gel -> new PCR 2b | ||
| + | |||
| + | <font color="#009933">PCR 2b</font> | ||
| + | |||
| + | - start PCR 2b with PCR 2b from 8-13-10 as template ( 1:20 and 1:100 diluted; 1µl) | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | - annealing temperature: 50°C; amount of water: 37,5µl | ||
| + | |||
| + | <font color="#009933">Agarose gel electrophoresis of PCR 2b 1:20 and 1:100</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | - expected bands: each ~ 700bp | ||
| + | |||
| + | - false bands: ~ 200bp | ||
| + | |||
| + | -> new PCR with 2ng, 5ng, 10ng template pDS7 | ||
| + | |||
| + | |||
| + | <font color="#009933">PCR 2b with 2ng, 5ng, 10ng template pDS7</font> | ||
| + | |||
| + | - pDS7 1:100 diluted(-> 2,1 ng/µl) | ||
| + | |||
| + | Mixture: | ||
| + | {| | ||
| + | |- | ||
| + | | | ||
| + | |2ng | ||
| + | |5ng | ||
| + | |10ng | ||
| + | |||
| + | |- | ||
| + | |Primer | ||
| + | |2*2,5µl (P5+P6) | ||
| + | |2*2,5µl (P5+P6) | ||
| + | |2*2,5µl (P5+P6) | ||
| + | |||
| + | |- | ||
| + | |10x Buffer Pfu | ||
| + | |5µl | ||
| + | |5µl | ||
| + | |5µl | ||
| + | |||
| + | |- | ||
| + | |dNTP Mix | ||
| + | |1µl | ||
| + | |1µl | ||
| + | |1µl | ||
| + | |||
| + | |- | ||
| + | |template | ||
| + | |pDS7 (dil.) | ||
| + | |1µl | ||
| + | |2,5µl | ||
| + | |5µl | ||
| + | |||
| + | |- | ||
| + | |Pfu Polymerase (3u/µl) | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |||
| + | |- | ||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |1,25µl | ||
| + | |1,25µl | ||
| + | |||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |33,25µl | ||
| + | |30,25µl | ||
| + | |25,25µl | ||
| + | |||
| + | |- | ||
| + | |sum | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |||
| + | |} | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | <font color="#009933">PCR 2a gel extraction</font> | ||
| + | |||
| + | - Quiagen kit (QuiaexII) | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/14 QIAEX II gel extraction| 14 QIAEX II gel extraction]] | ||
| + | |||
| + | <font color="#009933">Start 3 CMV overnight cultures</font> | ||
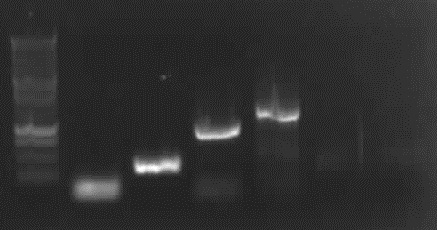
== 8-24-2010 == | == 8-24-2010 == | ||
| - | + | ||
| + | <font color="#009933">agarose gel electrophoresis of PCR 2b</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | [[Image:24_8_10_apo3.jpg|thumb|right|Agarose gel electrophoresis of (from left to right) PCR2b (2ng (cut out), 10ng, 5ng template) showing the right bands for 2ng, 5ng template]] | ||
| + | |||
| + | - expected bands:<font color="#CC33CC"> '''right bands with 2ng and 5ng template (~700bp)'''</font>, no band with 10ng template | ||
| + | |||
| + | <font color="#009933">CMV plasmid extraction</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/4 Plasmid extraction from cells|4 Plasmid extraction from cells]] | ||
| + | |||
| + | Plasmid extractionof 3 different overnight cultures. | ||
| + | |||
| + | - results: | ||
| + | * 52,5 ng/µl A260/A280= 1.312 | ||
| + | * 133 ng/µl A260/A280= 1.710 | ||
| + | * 80 ng/µl A260/A280= 1.600 | ||
| + | |||
| + | <font color="#009933">CMV restriction digestion</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/5 Restriction digest|5 Restriction digestion]] | ||
| + | |||
| + | - CMV restriction digest: EcoRI, PstI, buffer H | ||
| + | * 19µl, H<sub>2</sub>O : 0µl | ||
| + | * 6µl, H<sub>2</sub>O : 9.5µl | ||
| + | * 12.5µl, H<sub>2</sub>O : 3µl | ||
| + | |||
| + | <font color="#009933">PCR 2b gel extraction</font> | ||
| + | |||
| + | - PCR2b was gel extracted (with Qiagen gel extraction kit), 17.5 ng/µl a260/A280= 1.750 | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/14 QIAEX II gel extraction| 14 QIAEX II gel extraction]] | ||
| + | |||
| + | <font color="#009933">PCR 3 (fusion of 2a and 2b)</font> | ||
| + | |||
| + | - PCR3: conducted again at 52°C annealing temperature | ||
| + | |||
| + | * 10.5 ng (from PCR2b) 0.6µl | ||
| + | |||
| + | * 4.5 ng (from PCR2a) 0.9µl (1:10 diluted) | ||
| + | |||
| + | ::{| | ||
| + | |- | ||
| + | |PCR2a | ||
| + | |0.9 µl | ||
| + | |- | ||
| + | |PCR2b | ||
| + | |0.6 µl | ||
| + | |- | ||
| + | |primer3 | ||
| + | |2.5 µl | ||
| + | |- | ||
| + | |primer6 | ||
| + | |2.5 µl | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |1 µl | ||
| + | |- | ||
| + | |Pfu | ||
| + | |0.5 µl | ||
| + | |- | ||
| + | |10xbuffer | ||
| + | |5 µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |37 µl | ||
| + | |} | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | <font color="#009933">agarose gel electrophoresis of CMV digestion</font> | ||
| + | |||
| + | - agarose gel electrophoresis (150V, 25 min) of the CMV digestion | ||
| + | |||
| + | -> bands are wrong again ( ~ 1200bp, 2000bp) | ||
| + | |||
== 8-25-2010 == | == 8-25-2010 == | ||
| - | + | ||
| + | <font color="#009933">Agarose gel electrophorese of PCR 3</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | - expected band: ~1000bp | ||
| + | |||
| + | - false band: ~400bp | ||
| + | |||
| + | <font color="#009933">Plasmid extraction of ccdB tet and ccdB strep</font> | ||
| + | |||
| + | Plasmid extraction of pSB1C3 with BBa_P1010 | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/4 Plasmid extraction from cells|4 Plasmid extraction from cells]] | ||
| + | |||
| + | - results: | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |ccdB tet: | ||
| + | |50ng/µl; | ||
| + | |A260/A280= 1,818 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | <font color="#009933">Plate ccdB amp, cam, tet</font> | ||
| + | |||
| + | - plate ccdB with ampicilline, chloramphenicol, tetracycline resistance on LB agar with appropiate antibiotic. | ||
| + | |||
| + | |||
| + | <font color="#009933">Overnight culture of ccdB kan</font> | ||
| + | |||
| + | - Overnight culture of ccdB with kanamycine resistance in LB medium with kanamycine | ||
| + | |||
| + | <font color="#009933">PCR 7a, 7b, 9, 10</font> | ||
| + | |||
| + | ->Protocol: [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |||
| + | |PCR nr. | ||
| + | |template | ||
| + | |concentration | ||
| + | |dilution | ||
| + | |primer | ||
| + | |- | ||
| + | |||
| + | |7a | ||
| + | |190-6 | ||
| + | |~200ng/µl | ||
| + | |1:100 | ||
| + | |13,14 | ||
| + | |- | ||
| + | |||
| + | |7b | ||
| + | |190-6 | ||
| + | |~200ng/µl | ||
| + | |1:100 | ||
| + | |15,16 | ||
| + | |- | ||
| + | |||
| + | |9 | ||
| + | |eGFP | ||
| + | |55ng/µl | ||
| + | |1:25 | ||
| + | |20,21 | ||
| + | |- | ||
| + | |||
| + | |10 | ||
| + | |PhiC31o | ||
| + | |20ng/µl | ||
| + | |1:10 | ||
| + | |22,23 | ||
| + | |- | ||
| + | |||
| + | |} | ||
| + | |||
| + | |||
| + | Mixture | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |||
| + | |template (~2ng) | ||
| + | |1µl | ||
| + | |- | ||
| + | |||
| + | |Pfu | ||
| + | |0,5µl | ||
| + | |- | ||
| + | |||
| + | |Primer *2 | ||
| + | |2,5µl *2 | ||
| + | |- | ||
| + | |||
| + | |10x buffer | ||
| + | |5µl | ||
| + | |- | ||
| + | |||
| + | |dNTP Mix | ||
| + | |1µl | ||
| + | |- | ||
| + | |||
| + | |H<sub>2</sub>O | ||
| + | |37,5µl | ||
| + | |- | ||
| + | |||
| + | |sum | ||
| + | |50µl | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | Standard PCR; annealing temperature: 60°C | ||
| + | |||
== 8-26-2010 == | == 8-26-2010 == | ||
| - | + | <font color="#009933">Agarose gelelectrophoresis of PCR 7a, 7b, 9, 10</font> | |
| + | |||
| + | ->Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | - 150V, 25min | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |||
| + | |PCR nr. | ||
| + | |expected bands | ||
| + | |result | ||
| + | |- | ||
| + | |||
| + | |7a | ||
| + | |850bp | ||
| + | |no band | ||
| + | |- | ||
| + | |||
| + | |7b | ||
| + | |402bp | ||
| + | |false band (200bp) | ||
| + | |- | ||
| + | |||
| + | |9 | ||
| + | |808bp | ||
| + | |no band | ||
| + | |- | ||
| + | |||
| + | |10 | ||
| + | |1888bp | ||
| + | |no band | ||
| + | |- | ||
| + | |||
| + | |} | ||
| + | |||
| + | <font color="#009933">Plasmid extraction of ccdB kan</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/4 Plasmid extraction from cells|4 Plasmid extraction from cells]] | ||
| + | |||
| + | -result: concentration: 25ng/µl; A260/A280= 2,0 | ||
| + | |||
| + | <font color="#009933">New PCR 7a, 7b, 9, 10</font> | ||
| + | |||
| + | Mixture | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |||
| + | |template (~2ng) | ||
| + | |1µl | ||
| + | |- | ||
| + | |||
| + | |Pfu | ||
| + | |0,5µl | ||
| + | |- | ||
| + | |||
| + | |Primer *2 | ||
| + | |2,5µl *2 | ||
| + | |- | ||
| + | |||
| + | |10x buffer | ||
| + | |5µl | ||
| + | |- | ||
| + | |||
| + | |dNTP Mix | ||
| + | |1µl | ||
| + | |- | ||
| + | |||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |- | ||
| + | |||
| + | |H<sub>2</sub>O | ||
| + | |36,25µl | ||
| + | |- | ||
| + | |||
| + | |sum | ||
| + | |50µl | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | Program: | ||
| + | |||
| + | gradient PCR, 42-69°C annealing temp. | ||
| + | |||
| + | ->Protocol:[[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | <font color="#009933">Overnight culture of ccdB amp, tet, cam</font> | ||
| + | |||
| + | Inoculate one colony each in 5ml medium with approptraite antibiotic. | ||
| + | |||
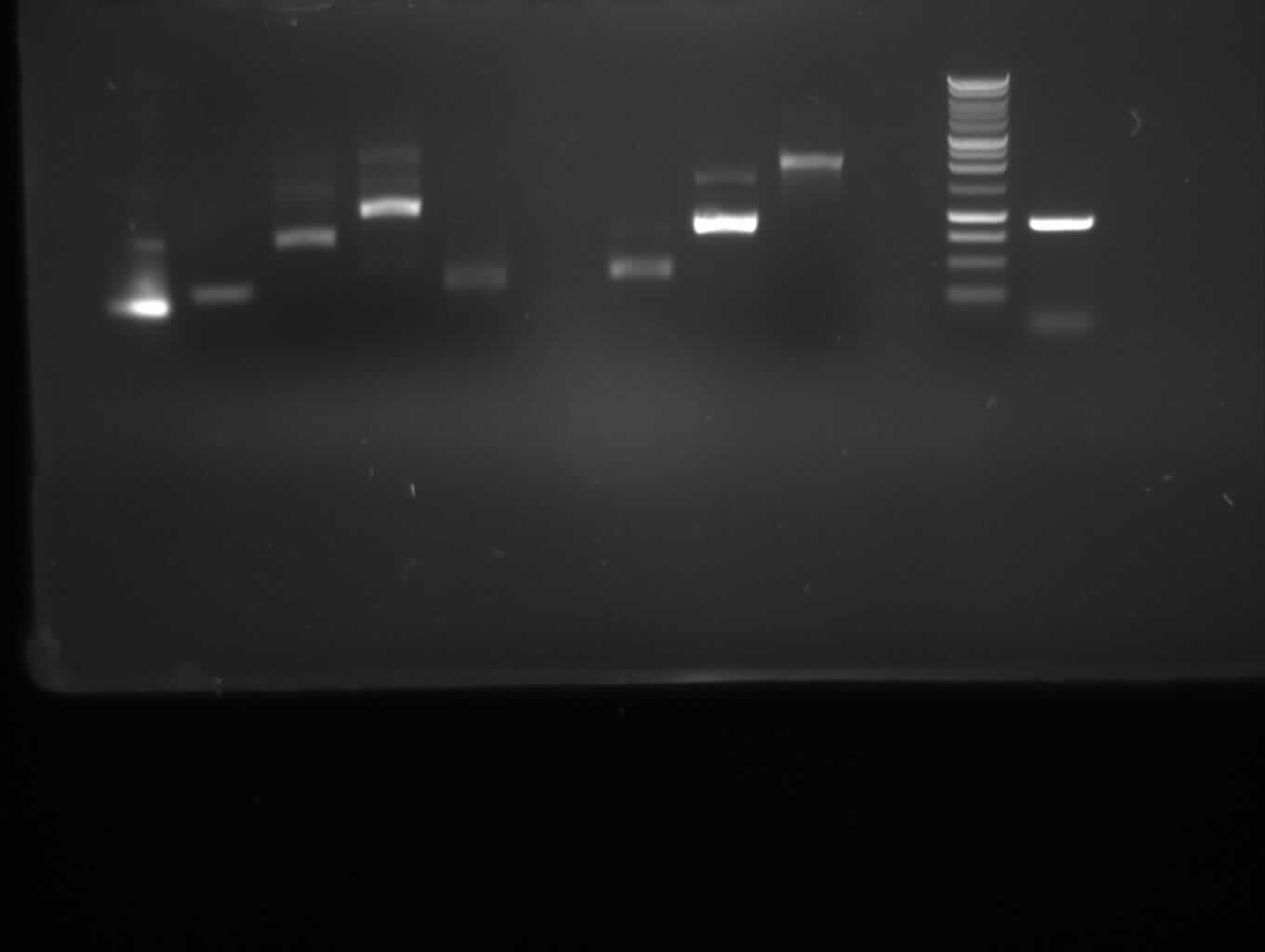
== 8-27-2010 == | == 8-27-2010 == | ||
| - | + | ||
| + | <font color="#009933">Agarose gel electrophoresis of PCR 7a, 7b, 9, 10</font> | ||
| + | |||
| + | ->Protocol:[[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | 150V, 25min, 75mA | ||
| + | |||
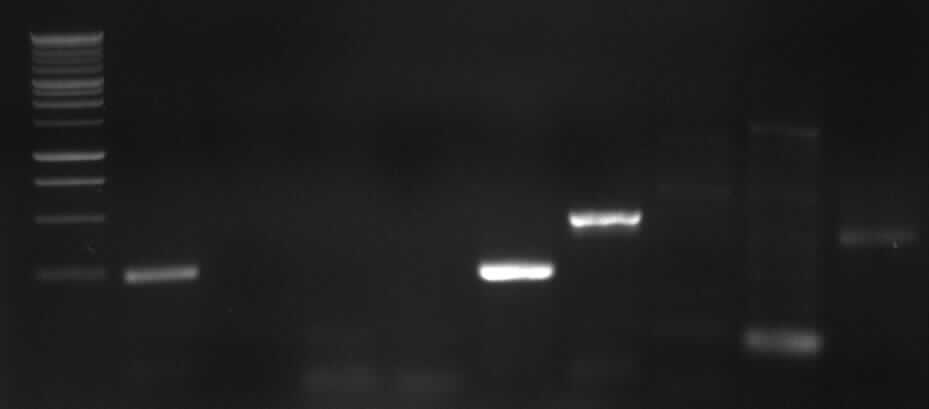
| + | [[Image: 27-8-10-1.jpg|400px|gel electrophoresis of new PCR 7a,7b, 9, 10]] | ||
| + | |||
| + | from left to right: 7a, 7b, 9, 10, Marker | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |||
| + | |PCR nr. | ||
| + | |expected bands | ||
| + | | result | ||
| + | |- | ||
| + | |||
| + | |7a | ||
| + | |850bp | ||
| + | |no band | ||
| + | |- | ||
| + | |||
| + | |<font color="#CC33CC">'''7b'''</font> | ||
| + | |<font color="#CC33CC">'''402bp'''</font> | ||
| + | |<font color="#CC33CC">'''right band (~400bp)'''</font>+ false band (~150bp) | ||
| + | |- | ||
| + | |||
| + | |9 | ||
| + | |808bp | ||
| + | |false band (~200bp) | ||
| + | |- | ||
| + | |||
| + | |<font color="#CC33CC">'''10'''</font> | ||
| + | |<font color="#CC33CC">'''1888bp'''</font> | ||
| + | |<font color="#CC33CC">'''right band (~1900bp)'''</font>+false band (~500bp) | ||
| + | |- | ||
| + | |||
| + | |} | ||
| + | |||
| + | <font color="#009933">Plasmid extraktion of ccdB amp, tet, cam</font> | ||
| + | |||
| + | ->Protocol: [[Team:LMU-Munich/Notebook/Protocols/4 Plasmid extraction from cells|4 Plasmid extraction from cells]] | ||
| + | |||
| + | results: | ||
| + | |||
| + | {| | ||
| + | |||
| + | |Plasmid | ||
| + | |concentration | ||
| + | |A260/A280 | ||
| + | |- | ||
| + | |||
| + | |ccdB amp | ||
| + | |57,5 ng/µl | ||
| + | |1,917 | ||
| + | |- | ||
| + | |||
| + | |||
| + | |ccdB cam | ||
| + | |70,0 ng/µl | ||
| + | |1,867 | ||
| + | |- | ||
| + | |||
| + | |||
| + | |ccdB tet | ||
| + | |50,0 ng/µl | ||
| + | |1,818 | ||
| + | |- | ||
| + | |||
| + | |} | ||
| + | |||
| + | |||
| + | <font color="#009933">New PCR 7a, 9</font> | ||
| + | |||
| + | Mixture: | ||
| + | |||
| + | - 2ng template: see 26-8-10 | ||
| + | |||
| + | - 4ng template: see 26-8-10, but 2µl template and 35,25µl water | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | |||
| + | <font color="#009933">Restriction digestion of ccdB amp, kan, cam, tet</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/5 Restriction digest|5 Restriction digestion]] | ||
| + | |||
| + | - only 90min 37°C incubation | ||
| + | |||
| + | - EcoRI, PstI, Buffer H | ||
| + | |||
| + | {| | ||
| + | |||
| + | |template | ||
| + | |volume | ||
| + | |mass | ||
| + | |- | ||
| + | |||
| + | |ccdB amp | ||
| + | |16µl | ||
| + | |930ng | ||
| + | |- | ||
| + | |||
| + | |ccdB cam | ||
| + | |14,3µl | ||
| + | |1µg | ||
| + | |- | ||
| + | |||
| + | |ccdB tet | ||
| + | |16µl | ||
| + | |800ng | ||
| + | |- | ||
| + | |||
| + | |ccdB kan | ||
| + | |16µl | ||
| + | |400ng | ||
| + | |- | ||
| + | |||
| + | |} | ||
| + | |||
| + | <font color="#009933">Agarose gelelectrophoresis of PCR 7a, 9, ccdB restriction digestion</font> | ||
| + | |||
| + | 150v, 25min, 75mA | ||
| + | |||
| + | -> Protocol:[[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | [[Image: 27 8 10 ccdb.jpg|400px|gel electrophoresis of new PCR 7a, 9, ccdb]] | ||
| + | |||
| + | results: | ||
| + | |||
| + | - PCR7a, 9: false band at 200bp | ||
| + | |||
| + | - <font color="#CC33CC">'''ccdB: each digestion leads to a right band with ~ 650bp'''</font> | ||
| + | |||
== 8-28-2010 == | == 8-28-2010 == | ||
weekend | weekend | ||
| Line 612: | Line 1,192: | ||
== 8-30-2010 == | == 8-30-2010 == | ||
| - | + | ||
| + | |||
| + | <font color="#009933">New PCR 7a and 9</font> | ||
| + | |||
| + | |||
| + | Mixture | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |||
| + | |template (~4ng) | ||
| + | |2µl | ||
| + | |- | ||
| + | |||
| + | |Pfu | ||
| + | |0,5µl | ||
| + | |- | ||
| + | |||
| + | |Primer *2 | ||
| + | |2,5µl *2 | ||
| + | |- | ||
| + | |||
| + | |10x buffer | ||
| + | |5µl | ||
| + | |- | ||
| + | |||
| + | |dNTP Mix | ||
| + | |1µl | ||
| + | |- | ||
| + | |||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |- | ||
| + | |||
| + | |H<sub>2</sub>O | ||
| + | |35,25µl | ||
| + | |- | ||
| + | |||
| + | |sum | ||
| + | |50µl | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | |||
| + | PCR program | ||
| + | |||
| + | PCR 7: Annealing Temperature 60°C - 25 x 1 min Annealing time and 5x 1,30 min Annealing time | ||
| + | |||
| + | PCR 9: Annealing Temperature 55°C - 25 x 1 min Annealing time and 5x 1,30 min Annealing time | ||
| + | |||
| + | |||
| + | <font color="#009933">Gel extraction of PCR 7b, 10</font> | ||
| + | |||
| + | ->Protocol: [[Team:LMU-Munich/Notebook/Protocols/14 QIAEX II gel extraction| 14 QIAEX II gel extraction]] | ||
| + | |||
| + | results: | ||
| + | |||
| + | {| | ||
| + | |||
| + | |PCR nr. | ||
| + | |concentration | ||
| + | |A260/A280 | ||
| + | |- | ||
| + | |||
| + | |7b | ||
| + | |10 ng/µl | ||
| + | |2,0 | ||
| + | |- | ||
| + | |||
| + | |||
| + | |10 | ||
| + | |17,5 ng/µl | ||
| + | |1,4 | ||
| + | |- | ||
| + | |||
| + | |} | ||
| + | |||
| + | |||
| + | <font color="#009933">Agarose gel electrophoresis of new PCR 7a, 9</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | 150V, 25min | ||
| + | |||
| + | - results: | ||
| + | |||
| + | - 7a: no band shown | ||
| + | |||
| + | - 9: false band (~200bp) | ||
| + | |||
| + | <font color="#009933">New PCR 3</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | ::{| | ||
| + | |- | ||
| + | |PCR2a | ||
| + | |0.9 µl | ||
| + | |- | ||
| + | |PCR2b | ||
| + | |0.6 µl | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |1 µl | ||
| + | |- | ||
| + | |Pfu | ||
| + | |0.5 µl | ||
| + | |- | ||
| + | |10xbuffer | ||
| + | |5 µl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |36,75 µl | ||
| + | |- | ||
| + | |sum | ||
| + | |45µl | ||
| + | |} | ||
| + | |||
| + | - new method: standard PCR without primers (10 cycles, 56°C annealing temp.) | ||
| + | |||
| + | - then add 2,5µl of primer 3 and 6 | ||
| + | |||
| + | - 30 cycles standard PCR (54°C annealing temp.) | ||
| + | |||
== 8-31-2010 == | == 8-31-2010 == | ||
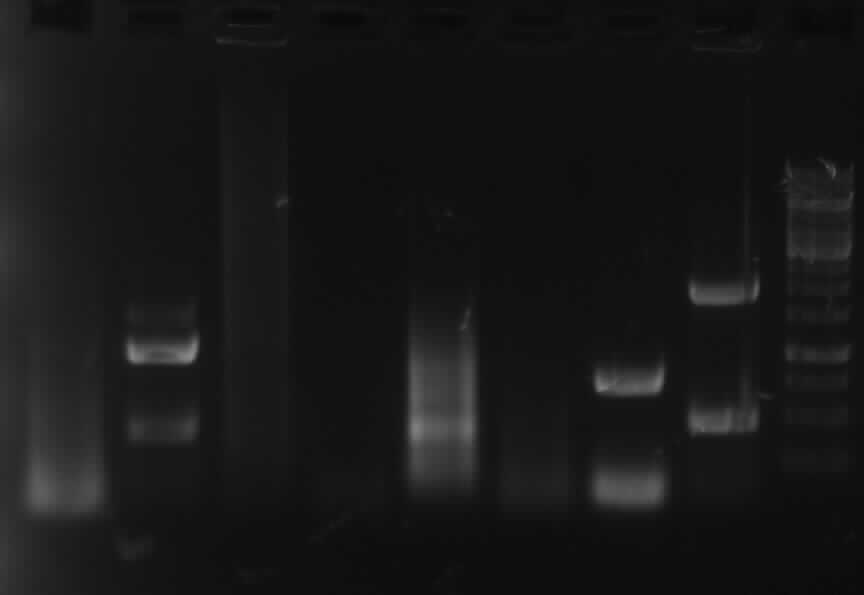
| - | + | [[Image:31 8 10 apo1.jpg|thumb|right|Gel photo of (left to right) PCR4a(2.5ng template), PCR4a(5ng template),PCR4b(2.5ng template), PCR4b(5ng template), PCR3(Pfu), PCR3(Phusion)]] | |
| + | |||
| + | <font color="#009933">Agarose gel electrophoresis of PCR 4a, PCR4b, PCR3(Pfu), PCR3(Phusion)</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | 150V, 25min | ||
| + | |||
| + | - results: | ||
| + | |||
| + | - PCR4a(2.5ng template), PCR4a(5ng template),PCR4b(2.5ng template), PCR4b(5ng template), PCR3(Pfu): no band shown | ||
| + | |||
| + | - <font color="#CC33CC">'''PCR3 (Phusion): right band (~1000bp)'''</font> | ||
| + | |||
| + | <font color="#009933">New PCR PCR4a, PCR4b, PCR7a, PCR9</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | PCR mixture for PCR4a, PCR4b | ||
| + | ::{| | ||
| + | |- | ||
| + | |template | ||
| + | |37.25 µl (200ng) | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |1 µl | ||
| + | |- | ||
| + | |Pfu | ||
| + | |0.5 µl | ||
| + | |- | ||
| + | |10xbuffer | ||
| + | |5 µl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |- | ||
| + | |sum | ||
| + | |50µl | ||
| + | |} | ||
| + | |||
| + | Standard PCR program with annealing temperature PCR4a: 51.1°C, PCR4b: 48.5°C. | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/15 PCR with Phusion| 15 PCR with Phusion]] | ||
| + | |||
| + | PCR mixture for PCR7a, PCR9 | ||
| + | ::{| | ||
| + | |- | ||
| + | |template | ||
| + | |4 µl (8ng) | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |1 µl | ||
| + | |- | ||
| + | |Phusion | ||
| + | |0.5 µl | ||
| + | |- | ||
| + | |5xbuffer | ||
| + | |10 µl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |28.25 µl | ||
| + | |- | ||
| + | |sum | ||
| + | |50µl | ||
| + | |} | ||
| + | |||
| + | PCR program: Phu62 | ||
| + | ::{| | ||
| + | |- | ||
| + | |98°C | ||
| + | |1 min | ||
| + | |- | ||
| + | |98°C | ||
| + | |10 sec | ||
| + | |- | ||
| + | |62°C | ||
| + | |20 sec | ||
| + | |- | ||
| + | |73°C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |return to step 2 for 29 cycles | ||
| + | | | ||
| + | |- | ||
| + | |73°C | ||
| + | |10 min | ||
| + | |- | ||
| + | |12°C | ||
| + | |forever | ||
| + | |} | ||
| + | |||
| + | |||
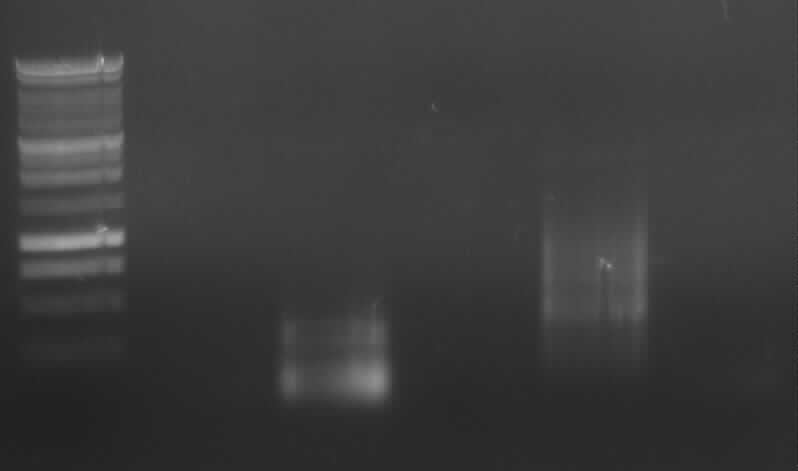
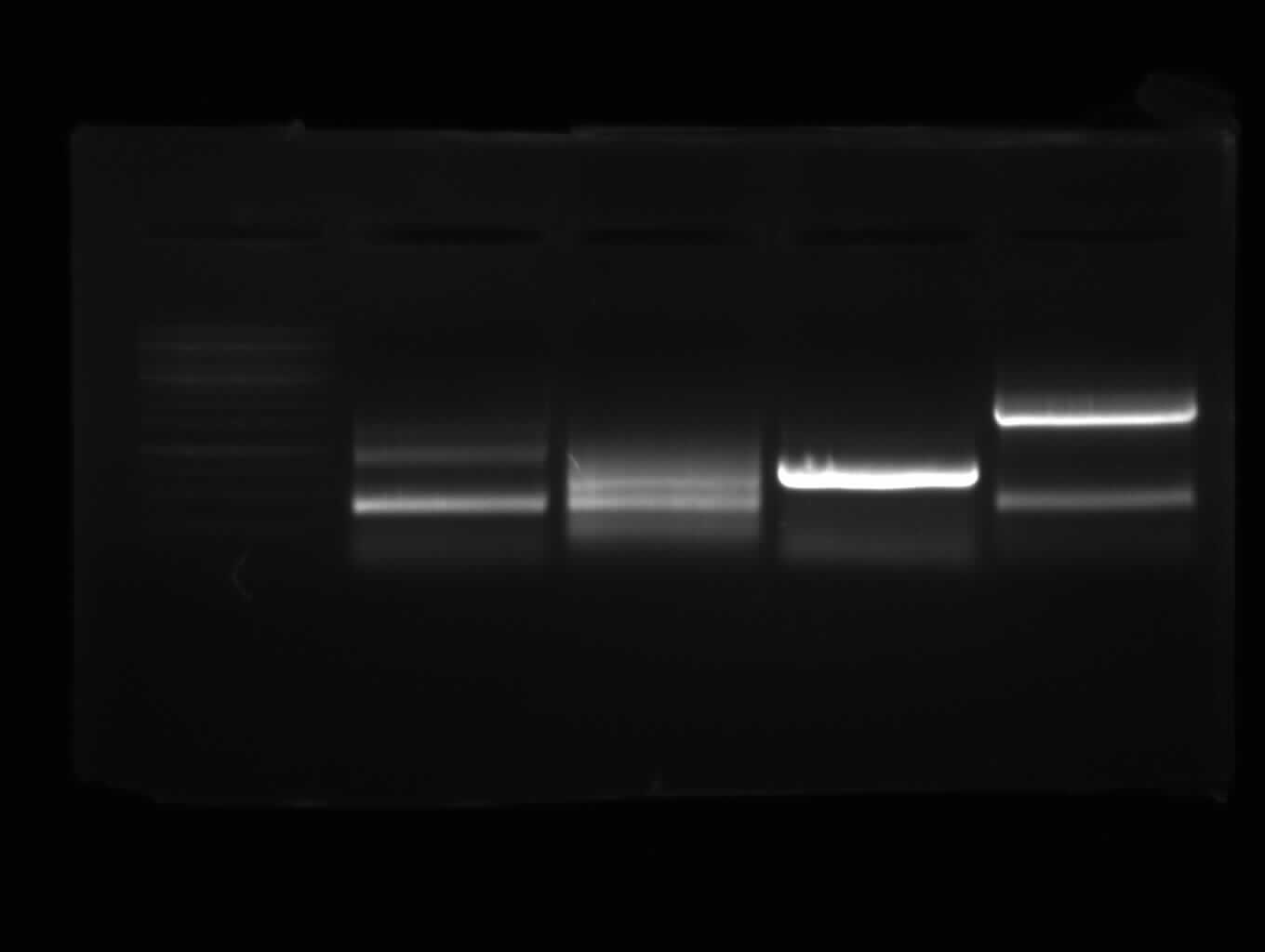
| + | [[Image:31 8 10 apo2.jpg|thumb|right|Gel photo of (left to right) PCR3, ladder, PCR7a, empty, PCR9]] | ||
| + | |||
| + | <font color="#009933">Agarose gel electrophoresis of PCR 3, PCR7a, PCR9 for gel extraction</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | 120V, 30min | ||
| + | |||
| + | - results: | ||
| + | |||
| + | -<font color="#CC33CC"> '''PCR3; right band (~1000bp)'''</font> and side-product | ||
| + | |||
| + | - PCR7a: no band | ||
| + | |||
| + | - PCR9: right band (~800bp) and side-product | ||
| + | |||
| + | <font color="#009933">Gel extraction of the DNA from PCR3 and PCR9</font> | ||
| + | |||
| + | -> Protocol ([[Team:LMU-Munich/Notebook/Protocols/12_Gel_extraction_or_PCR_Clean_up|12 Gel extraction or PCR Clean up]]) | ||
| + | |||
| + | results: | ||
| + | |||
| + | PCR 9: 22,5ng/µl; A260/A280=1,8 | ||
| + | |||
| + | PCR 3: 22,5ng/µl; A260/A280=2,25 | ||
| + | |||
| + | <font color="#009933">New PCR 4a, 4b, 7a with DreamTaq</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/16 PCR with DreamTaq| 16 PCR with DreamTaq]] | ||
| + | |||
| + | PCR mixture for PCR7a | ||
| + | ::{| | ||
| + | |- | ||
| + | |template | ||
| + | |5 µl (10ng) | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |5 µl | ||
| + | |- | ||
| + | |DreamTaq | ||
| + | |0.33 µl | ||
| + | |- | ||
| + | |10xbuffer | ||
| + | |5 µl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |28.5 µl | ||
| + | |- | ||
| + | |sum | ||
| + | |50µl | ||
| + | |} | ||
| + | |||
| + | Primers for PCR 7a: 13,14 | ||
| + | |||
| + | Annealing temp: 60°C | ||
| + | |||
| + | PCR mixture for PCR4a,4b | ||
| + | ::{| | ||
| + | |- | ||
| + | |template | ||
| + | |36,5 µl (180ng HeLa cDNA) | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |5 µl | ||
| + | |- | ||
| + | |DreamTaq | ||
| + | |0.33 µl | ||
| + | |- | ||
| + | |10xbuffer | ||
| + | |5 µl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |- | ||
| + | |sum | ||
| + | |50µl | ||
| + | |} | ||
| + | |||
| + | PCR 4a | ||
| + | |||
| + | Primers for PCR 4a: 7,8 | ||
| + | |||
| + | Primers for PCR 4b: 9,10 | ||
| + | |||
| + | Annealing temp: 50°C | ||
| + | |||
| + | PCR program: | ||
| + | ::{| | ||
| + | |- | ||
| + | |95°C | ||
| + | |1 min | ||
| + | |- | ||
| + | |95°C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |50/60°C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |72°C | ||
| + | |1 min (1kb/min) | ||
| + | |- | ||
| + | |return to step 2 for 29 cycles | ||
| + | | | ||
| + | |- | ||
| + | |72°C | ||
| + | |10 min | ||
| + | |- | ||
| + | |12°C | ||
| + | |forever | ||
| + | |} | ||
| + | |||
== 9-01-2010 == | == 9-01-2010 == | ||
| - | + | ||
| + | <font color="#009933">Agarose gel electrophoresis of PCR4a, PCR4b, PCR7 (DreamTaq)</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | 150V, 25min | ||
| + | |||
| + | results: no product | ||
| + | |||
| + | <font color="#009933">New PCR 4a, 4b with DreamTaq, Pfu, with concentration gradient and touch-down PCR </font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/16 PCR with DreamTaq| 16 PCR with DreamTaq]]; [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | PCR mixture for DreamTaq | ||
| + | ::{| | ||
| + | |- | ||
| + | |Concentration | ||
| + | |Low | ||
| + | |Middle | ||
| + | |High | ||
| + | |- | ||
| + | |template | ||
| + | |5 µl (1:1000) | ||
| + | |31.75µl (1:1000) | ||
| + | |2µl (1:10) | ||
| + | |- | ||
| + | |dNTPs | ||
| + | | | ||
| + | |5 µl | ||
| + | | | ||
| + | |- | ||
| + | |DreamTaq | ||
| + | | | ||
| + | |0.33 µl | ||
| + | | | ||
| + | |- | ||
| + | |10xbuffer | ||
| + | | | ||
| + | |5 µl | ||
| + | | | ||
| + | |- | ||
| + | |DMSO | ||
| + | | | ||
| + | |1,25µl | ||
| + | | | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |26.75 µl | ||
| + | |0 | ||
| + | |29.75 | ||
| + | |- | ||
| + | |sum | ||
| + | | | ||
| + | |50µl | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | PCR mixture for Pfu | ||
| + | |||
| + | ::{| | ||
| + | |- | ||
| + | |Concentration | ||
| + | |Low | ||
| + | |Middle | ||
| + | |High | ||
| + | |- | ||
| + | |template | ||
| + | |5 µl (1:1000) | ||
| + | |37.25µl (1:1000) | ||
| + | |2µl (1:10) | ||
| + | |- | ||
| + | |dNTPs | ||
| + | | | ||
| + | |1 µl | ||
| + | | | ||
| + | |- | ||
| + | |Pfu | ||
| + | | | ||
| + | |0.5 µl | ||
| + | | | ||
| + | |- | ||
| + | |10xbuffer | ||
| + | | | ||
| + | |5 µl | ||
| + | | | ||
| + | |- | ||
| + | |DMSO | ||
| + | | | ||
| + | |1,25µl | ||
| + | | | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |32.25 µl | ||
| + | |0 | ||
| + | |35.25 µl | ||
| + | |- | ||
| + | |sum | ||
| + | | | ||
| + | |50µl | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | Primers for PCR 4a: 7,8; PCR 4b: 9,10 | ||
| + | |||
| + | -> Protocol: Thermal cycler program: [[Team:LMU-Munich/Notebook/Protocols/17 Special thermal cycler programs/Touch down|Touch down]] | ||
| + | |||
| + | <font color="#009933">Agarose gel electrophoresis of PCR4a, PCR4b</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | 150V, 25min | ||
| + | |||
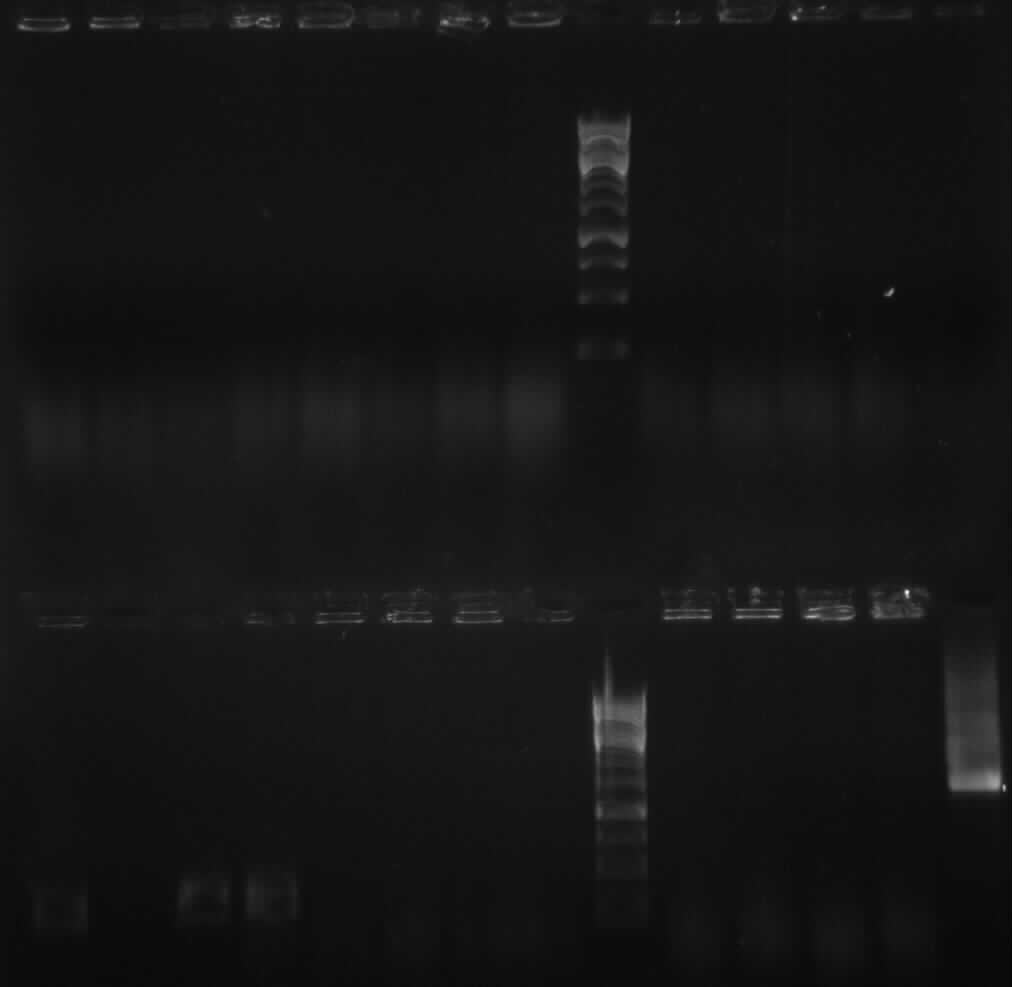
| + | [[image: 9.1.10apo.jpg|400px|gel 9-1-10]] | ||
| + | |||
| + | from left to right: 4a: P1, P2, P3, D1, D2, D3; 4b: P1, P2, P3, D1, D2, D3 | ||
| + | |||
| + | key: | ||
| + | |||
| + | "P"= PCR with Pfu | ||
| + | |||
| + | "D"= PCR with DreamTaq | ||
| + | |||
| + | "1"= low template concentration | ||
| + | |||
| + | "2"= middle template concentration | ||
| + | |||
| + | "3"= high template concentration | ||
| + | |||
| + | expected bands: | ||
| + | |||
| + | *<font color="#CC33CC">'''4a: 330bp -> P2 and P3 show right bands '''</font>and "primer clouds"(?) | ||
| + | |||
| + | *<font color="#CC33CC">'''4b: 376bp -> P1, P2, P3 show right bands'''</font> and "primer clouds" (?) | ||
| + | |||
== 9-02-2010 == | == 9-02-2010 == | ||
| - | + | <font color="#009933">Agarose gel electrophorese of PCR 4a P2, 4b P2</font> | |
| - | == 9- | + | |
| - | + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | |
| + | |||
| + | - 120V, 45min, 1,5% Agarose gel | ||
| + | |||
| + | [[Image:2 9 10 apo.jpg|400px|Agarose gel electrophoresis of (from left to right) PCR4aP2, Marker and PCR4bP2]] | ||
| + | |||
| + | Agarose gel electrophoresis of (from left to right) PCR4aP2, Marker and PCR4bP2 | ||
| + | |||
| + | |||
| + | - Cut out bands at ~~ 350bp and extract | ||
| + | |||
| + | <font color="#009933">PCR Agarose gel extraction</font> | ||
| + | |||
| + | -> Protocol:[[Team:LMU-Munich/Notebook/Protocols/14 QIAEX II gel extraction| 14 QIAEX II gel extraction]] | ||
| + | |||
| + | results: | ||
| + | |||
| + | {| | ||
| + | |||
| + | |PCR nr. | ||
| + | |concentration | ||
| + | |A260/A280 | ||
| + | |- | ||
| + | |||
| + | |4aP2 | ||
| + | |12,5 ng/µl | ||
| + | |1,67 | ||
| + | |- | ||
| + | |||
| + | |||
| + | |4bP2 | ||
| + | |72,5 ng/µl | ||
| + | |1,53 | ||
| + | |- | ||
| + | |||
| + | |} | ||
| + | |||
| + | |||
| + | <font color="#009933">New PCR 7a with Pfu and Phusion</font> | ||
| + | |||
| + | template: 190-6, Primer 13,14 | ||
| + | |||
| + | Mixture with Pfu | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |||
| + | |template (~2ng) | ||
| + | |1µl | ||
| + | |- | ||
| + | |||
| + | |Pfu | ||
| + | |0,5µl | ||
| + | |- | ||
| + | |||
| + | |Primer *2 | ||
| + | |2,5µl *2 | ||
| + | |- | ||
| + | |||
| + | |10x buffer | ||
| + | |5µl | ||
| + | |- | ||
| + | |||
| + | |dNTP Mix | ||
| + | |1µl | ||
| + | |- | ||
| + | |||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |- | ||
| + | |||
| + | |H<sub>2</sub>O | ||
| + | |36,25µl | ||
| + | |- | ||
| + | |||
| + | |sum | ||
| + | |50µl | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | PCR mixture with Phusion | ||
| + | ::{| | ||
| + | |- | ||
| + | |template (~2ng) | ||
| + | |1,0 µl | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |1 µl | ||
| + | |- | ||
| + | |Phusion | ||
| + | |0.5 µl | ||
| + | |- | ||
| + | |5xbuffer | ||
| + | |10 µl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |31,25µl | ||
| + | |- | ||
| + | |sum | ||
| + | |50µl | ||
| + | |} | ||
| + | |||
| + | -> Protocol:[[Team:LMU-Munich/Notebook/Protocols/15 PCR with Phusion| 15 PCR with Phusion]] | ||
| + | |||
| + | <font color="#009933">New PCR 4a, 4b with Pfu</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | -> Protocol: Thermal cycler program: [[Team:LMU-Munich/Notebook/Protocols/17 Special thermal cycler programs/Touch down|Touch down]] | ||
| + | |||
| + | Mixture see 9-1-10, twice 4a and 4b | ||
| + | |||
| + | <font color="#009933">Agarose gel electrophorese of PCR 4a, 4b, 7a, gel extracted 4a and 4b</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | - 25min, 150V | ||
| + | |||
| + | [[image:9-2-10apo2.jpg|400px|gel 9-2-10-2]] | ||
| + | |||
| + | from left to right: 4a*, 4a, 4b*, 4b, 7a Phusion, 7a Pfu, Ladder | ||
| + | |||
| + | -> <font color="#CC33CC">'''result: 4b, 4b*: right bands (~330bp)'''</font> | ||
| + | |||
| + | -remain: false bands/no band | ||
| + | |||
| + | |||
| + | [[image:9-2-10apo3.jpg|400px|gel 9-2-10-3]] | ||
| + | |||
| + | from left to right: ladder, 4 columns pathway, 4a gelextr., 4b gelextr. | ||
| + | |||
| + | -> <font color="#CC33CC">'''results: slight right bands for 4a and 4b'''</font>, no "primer clouds" anymore. | ||
| + | |||
| + | == 9-03-2010 == | ||
| + | |||
| + | <font color="#009933">Overlapping PCR 5 with Pfu and Phusion</font> | ||
| + | |||
| + | template: 4a, 4b Primer 7,10 | ||
| + | |||
| + | Mixture with Pfu | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |PCR4a (~15ng) | ||
| + | |1.2µl | ||
| + | |- | ||
| + | |PCR4b (~15ng) | ||
| + | |2µl (1:10) | ||
| + | |- | ||
| + | |Pfu | ||
| + | |0.5µl | ||
| + | |- | ||
| + | |Primer *2 | ||
| + | |2.5µl *2 | ||
| + | |- | ||
| + | |10x buffer | ||
| + | |5µl | ||
| + | |- | ||
| + | |dNTP Mix | ||
| + | |1µl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |34.05µl | ||
| + | |- | ||
| + | |sum | ||
| + | |50µl | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | PCR program: standard PCR program for Pfu, Annealing temperature: 54°C | ||
| + | |||
| + | PCR mixture with Phusion | ||
| + | ::{| | ||
| + | |- | ||
| + | |PCR4a (~15ng) | ||
| + | |1.2µl | ||
| + | |- | ||
| + | |PCR4b (~15ng) | ||
| + | |2µl (1:10) | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |1 µl | ||
| + | |- | ||
| + | |Phusion | ||
| + | |0.5 µl | ||
| + | |- | ||
| + | |5xbuffer | ||
| + | |10 µl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |29.05µl | ||
| + | |- | ||
| + | |sum | ||
| + | |50µl | ||
| + | |} | ||
| + | |||
| + | -> Protocol:[[Team:LMU-Munich/Notebook/Protocols/15 PCR with Phusion| 15 PCR with Phusion]] | ||
| + | |||
| + | PCR program: standard PCR program for Phusion, Annealing temperature: 58°C | ||
| + | |||
| + | <font color="#009933">New PCR7a with Pfu</font> | ||
| + | |||
| + | template: 190-6; Primer: 13,14 | ||
| + | |||
| + | Mixture with Pfu | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |template (~2ng) | ||
| + | |1µl | ||
| + | |- | ||
| + | |Pfu | ||
| + | |0.5µl | ||
| + | |- | ||
| + | |Primer *2 | ||
| + | |2.5µl *2 | ||
| + | |- | ||
| + | |10x buffer | ||
| + | |5µl | ||
| + | |- | ||
| + | |dNTP Mix | ||
| + | |1µl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |36.25µl | ||
| + | |- | ||
| + | |sum | ||
| + | |50µl | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | PCR program: standard PCR program for Pfu, Gradient: 54.4°C, 57.8°C, 61.4°C, 65.0°C | ||
| + | |||
| + | -> results: | ||
| + | |||
| + | -7a: only "primer cluods" | ||
| + | |||
| + | -5 Pfu: no defined product (slurred?) | ||
| + | |||
| + | -5 Phusion: "primer clouds" | ||
| + | |||
== 9-04-2010 == | == 9-04-2010 == | ||
weekend | weekend | ||
| Line 627: | Line 1,936: | ||
== 9-06-2010 == | == 9-06-2010 == | ||
| - | + | charges for sequencing | |
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! name | ||
| + | ! 4a-7 | ||
| + | ! 4a-8 | ||
| + | ! 4b-9 | ||
| + | ! 4b-10 | ||
| + | ! 3-3 | ||
| + | ! 3-6 | ||
| + | ! 6-11 | ||
| + | ! 6-12 | ||
| + | |- | ||
| + | | DNA | ||
| + | | 4a; 2.4µl | ||
| + | | 4a; 2.4µl | ||
| + | | 4b; 0.5µl | ||
| + | | 4b; 0.5µl | ||
| + | | 3; 2µl | ||
| + | | 3; 2µl | ||
| + | | 6; 0.5µl | ||
| + | | 6; 0.5µl | ||
| + | |- | ||
| + | | primer [1pmol/µl] | ||
| + | | 7; 3.2µl | ||
| + | | 8; 3.2µl | ||
| + | | 9; 3.2µl | ||
| + | | 10; 3.2µl | ||
| + | | 3; 3.2µl | ||
| + | | 6; 3.2µl | ||
| + | | 11; 3.2µl | ||
| + | | 12; 3.2µl | ||
| + | |- | ||
| + | | Tris (10mM); pH 8.2 | ||
| + | | 1.4µl | ||
| + | | 1.4µl | ||
| + | | 3.3µl | ||
| + | | 3.3µl | ||
| + | | 1.8µl | ||
| + | | 1.8µl | ||
| + | | 3.3µl | ||
| + | | 3.3µl | ||
| + | |} | ||
| + | every charge 7µl | ||
| + | |||
| + | |||
| + | <font color="#009933">New overlapping PCR 5 with Pfu</font> | ||
| + | |||
| + | template: 4a, 4b; 15ng | ||
| + | |||
| + | Primer 7,10 | ||
| + | |||
| + | |||
| + | |||
| + | Mixture | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |PCR4a (~7ng) | ||
| + | |0.56µl | ||
| + | |- | ||
| + | |PCR4b (~8ng) | ||
| + | |1.1µl (1:10) | ||
| + | |- | ||
| + | |Pfu | ||
| + | |0.5µl | ||
| + | |- | ||
| + | |Primer *2 | ||
| + | |2.5µl *2 | ||
| + | |- | ||
| + | |10x buffer | ||
| + | |5µl | ||
| + | |- | ||
| + | |dNTP Mix | ||
| + | |1µl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |36.6µl | ||
| + | |- | ||
| + | |sum | ||
| + | |50µl | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | PCR program: standard PCR program for Pfu, Annealing temperature: 46.1°C,48.5°C, 51.1°C | ||
| + | |||
| + | -> results: "primer clouds" | ||
| + | |||
| + | |||
| + | <font color="#009933">New PCR 7a with Pfu</font> | ||
| + | new dilution of 190-6; 1:100 | ||
| + | |||
| + | {| | ||
| + | |template[2ng/µl] | ||
| + | |1µl | ||
| + | |2µl | ||
| + | |- | ||
| + | |pfu polymerase | ||
| + | |0.5µl | ||
| + | |0.5µl | ||
| + | |- | ||
| + | |primer13 | ||
| + | |2.5µl | ||
| + | |2.5µl | ||
| + | |- | ||
| + | |primer14 | ||
| + | |2.5µl | ||
| + | |2.5µl | ||
| + | |- | ||
| + | |buffer | ||
| + | |5µl | ||
| + | |5µl | ||
| + | |- | ||
| + | |dNTP Mix | ||
| + | |1µl | ||
| + | |1µl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |1.25µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |36.25µl | ||
| + | |35.25 | ||
| + | |- | ||
| + | |sum | ||
| + | |50µl | ||
| + | |50µl | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | <font color="#009933">Agarose gel electrophoresis of 7a</font> | ||
| + | |||
| + | -> results: "primer clouds" | ||
| + | |||
| + | |||
| + | <font color="#009933">charges for sequencing</font> for PCR 1,7b and 9 | ||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! name | ||
| + | ! 1-1 | ||
| + | ! 1-2 | ||
| + | ! 7b-15 | ||
| + | ! 7b-16 | ||
| + | ! 9-20 | ||
| + | ! 9-21 | ||
| + | ! 10-22 | ||
| + | ! 10-23 | ||
| + | |- | ||
| + | | DNA | ||
| + | | 1[1:10]; 1.74µl | ||
| + | | 1[1:10]; 1.74µl | ||
| + | | 7b; 3µl | ||
| + | | 7b; 3µl | ||
| + | | 9; 1.78µl | ||
| + | | 9; 1.78µl | ||
| + | | 10; 3.8µl | ||
| + | | 10; 3.8µl | ||
| + | |- | ||
| + | | primer [1pmol/µl] | ||
| + | | 1; 3.2µl | ||
| + | | 2; 3.2µl | ||
| + | | 15; 3.2µl | ||
| + | | 16; 3.2µl | ||
| + | | 20; 3.2µl | ||
| + | | 21; 3.2µl | ||
| + | | 22; 3.2µl | ||
| + | | 23; 3.2µl | ||
| + | |- | ||
| + | | Tris (10mM); pH 8.2 | ||
| + | | 2.06µl | ||
| + | | 2.06µl | ||
| + | | 0.8µl | ||
| + | | 0.8µl | ||
| + | | 2.02µl | ||
| + | | 2.02µl | ||
| + | | 0µl | ||
| + | | 0µl | ||
| + | |} | ||
| + | every charge 7µl | ||
| + | |||
== 9-07-2010 == | == 9-07-2010 == | ||
| - | + | <font color="#009933">Agarose gel electrophorese of PCR gel extractions</font> | |
| + | |||
| + | 150V, 25min | ||
| + | |||
| + | -> Protocol:[[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | -result: | ||
| + | |||
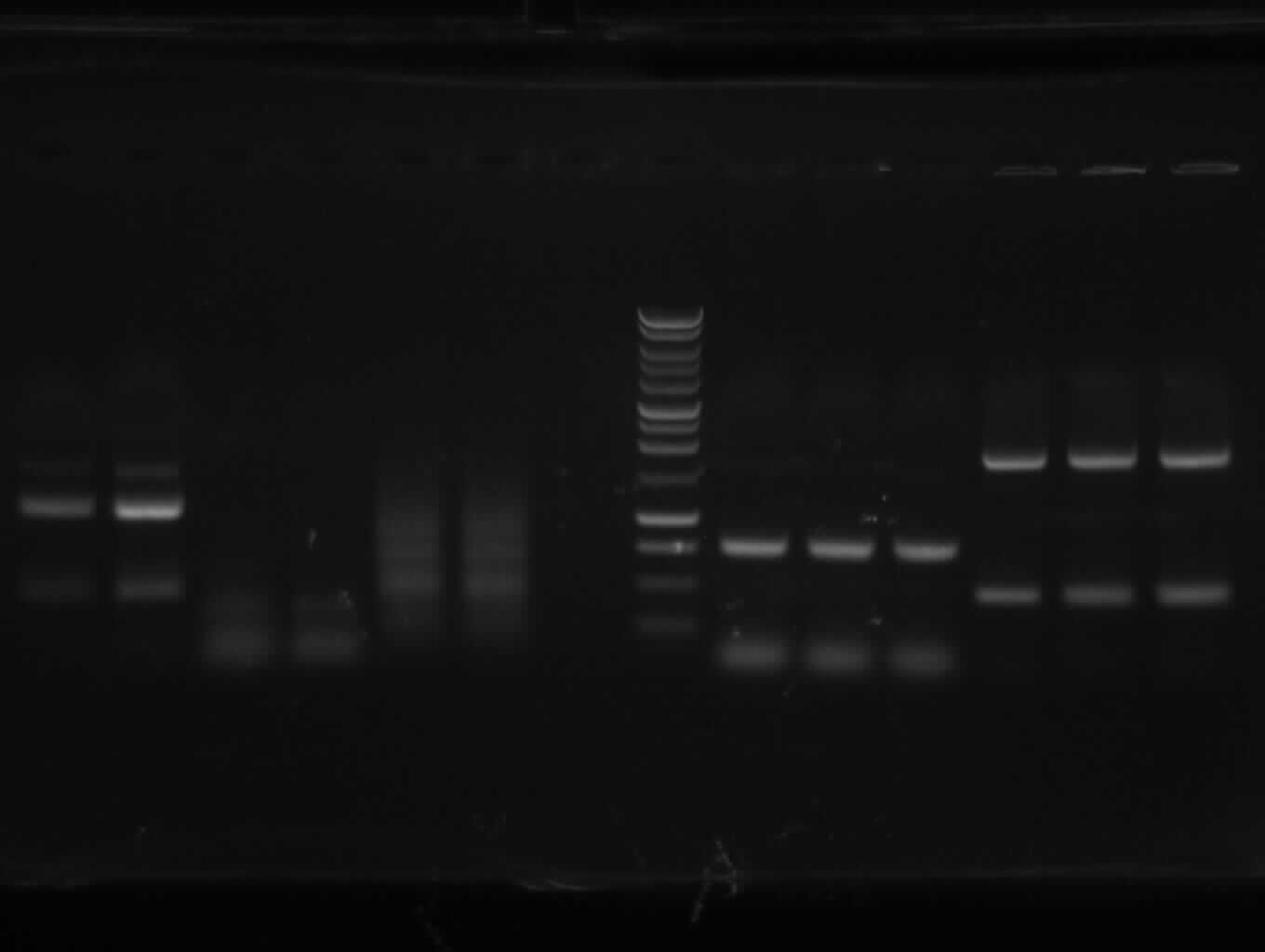
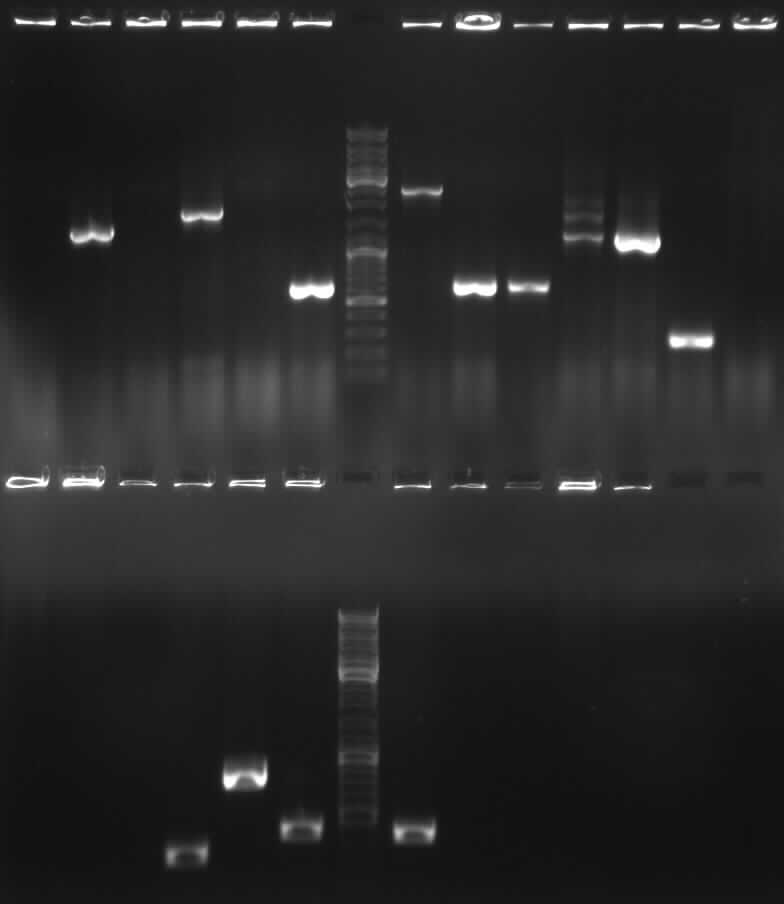
| + | [[image:7-9-10-1.jpg|400px|gel photo]] | ||
| + | |||
| + | from left to right: PCR 1,2a,2b,3,4a,/,7b,9,10,/,ladder,pathway | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | !PCR nr. | ||
| + | !1 | ||
| + | !<font color="#CC33CC">2a</font> | ||
| + | !<font color="#CC33CC">2b</font> | ||
| + | !<font color="#CC33CC">3</font> | ||
| + | !<font color="#CC33CC">4a</font> | ||
| + | !<font color="#CC33CC">7b</font> | ||
| + | !9 | ||
| + | !<font color="#CC33CC">10</font> | ||
| + | |- | ||
| + | |expected band (bp) | ||
| + | |492 | ||
| + | |332 | ||
| + | |772 | ||
| + | |1087 | ||
| + | |330 | ||
| + | |402 | ||
| + | |808 | ||
| + | |1888 | ||
| + | |- | ||
| + | |shown band(s) | ||
| + | |550,200 | ||
| + | |<font color="#CC33CC">'''300'''</font> | ||
| + | |<font color="#CC33CC">'''750'''</font> | ||
| + | |<font color="#CC33CC">'''1100'''</font> | ||
| + | |<font color="#CC33CC">'''300'''</font> | ||
| + | |<font color="#CC33CC">'''450'''</font> | ||
| + | |900,1500 | ||
| + | |<font color="#CC33CC">'''1900'''</font> | ||
| + | |- | ||
| + | |clean charge | ||
| + | | | ||
| + | |<font color="#CC33CC">'''x'''</font> | ||
| + | |<font color="#CC33CC">'''x'''</font> | ||
| + | |<font color="#CC33CC">'''~x'''</font> | ||
| + | |<font color="#CC33CC">'''x'''</font> | ||
| + | |<font color="#CC33CC">'''x'''</font> | ||
| + | | | ||
| + | |<font color="#CC33CC">'''x'''</font> | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | <font color="#009933">new PCR for PCR6</font> | ||
| + | |||
| + | PCR mixture for PCR6 | ||
| + | |||
| + | ::{| | ||
| + | |- | ||
| + | |Concentration | ||
| + | |Low | ||
| + | |High | ||
| + | |- | ||
| + | |template | ||
| + | |1 µl (1:100) | ||
| + | |15µl (1:100) | ||
| + | |- | ||
| + | |primer (11,12) | ||
| + | |2.5 µl*2 | ||
| + | | | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |1 µl | ||
| + | | | ||
| + | |- | ||
| + | |Pfu | ||
| + | |0.5 µl | ||
| + | | | ||
| + | |- | ||
| + | |10xbuffer | ||
| + | |5 µl | ||
| + | | | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | | | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |36.25 µl | ||
| + | |22.25 µl | ||
| + | |- | ||
| + | |sum | ||
| + | |50µl | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | -> Protocol: Thermal cycler program: [[Team:LMU-Munich/Notebook/Protocols/17 Special thermal cycler programs/Touch down|Touch down]], 62°C-52°C, 30 cycles by 55°C | ||
| + | |||
== 9-08-2010 == | == 9-08-2010 == | ||
| - | + | <font color="#009933">Restriction digestion of eGFP, PCR6, ccdBamp</font> | |
| + | |||
| + | [[Image:Apo_8.9.10.jpg|thumb|right|Gel photo of (left to right) PCR6(5ng/100ng) template]] | ||
| + | |||
| + | as prepatation for ligation | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/5_Restriction_digest|5 Restriction digestion]] | ||
| + | |||
| + | {| | ||
| + | !eGFP | ||
| + | ! | ||
| + | !SV40PA=PCR6 | ||
| + | ! | ||
| + | !ccdBamp | ||
| + | ! | ||
| + | |- | ||
| + | |DNA | ||
| + | |5µl=300ng | ||
| + | |DNA | ||
| + | |1µl=37,5ng | ||
| + | |DNA | ||
| + | |2µl=115ng | ||
| + | |- | ||
| + | |Buffer MC | ||
| + | |2µl | ||
| + | |Buffer D | ||
| + | |2µl | ||
| + | |||
| + | |Buffer H | ||
| + | |2µl | ||
| + | |- | ||
| + | |BSA 1:10 | ||
| + | |2µl | ||
| + | |BSA 1:10 | ||
| + | |2µl | ||
| + | |BSA 1:10 | ||
| + | |2µl | ||
| + | |- | ||
| + | |EcoRI | ||
| + | |0,5µl | ||
| + | |XbaI | ||
| + | |0,5µl | ||
| + | |EcoRI | ||
| + | |0,5µl | ||
| + | |- | ||
| + | |SpeI | ||
| + | |0,5µl | ||
| + | |PstI | ||
| + | |0,5µl | ||
| + | |PstI | ||
| + | |0,5µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |13µl | ||
| + | |H<sub>2</sub>O | ||
| + | |14µl | ||
| + | |H<sub>2</sub>O | ||
| + | |13µl | ||
| + | |- | ||
| + | |sum | ||
| + | |23µl | ||
| + | |sum | ||
| + | |20µl | ||
| + | |sum | ||
| + | |20µl | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | 1:30h 37°C, 20min 80°C | ||
| + | |||
| + | <font color="#009933">Agarose gel electrophoresis of PCR6</font> | ||
| + | |||
| + | -> Protocol:[[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | 25min, 150V | ||
| + | |||
| + | |||
| + | <font color="#009933">Ligation 1</font> | ||
| + | |||
| + | {| | ||
| + | |eGFP | ||
| + | |14,4µL | ||
| + | |(144ng) | ||
| + | |- | ||
| + | |SV40PA | ||
| + | |4,8µL | ||
| + | |(48ng) | ||
| + | |- | ||
| + | |ccdBamp | ||
| + | |2,9µL | ||
| + | |(100ng) | ||
| + | |- | ||
| + | |T4 Buffer 10x | ||
| + | |3µL | ||
| + | | | ||
| + | |- | ||
| + | |T4 Ligase | ||
| + | |0,5µL | ||
| + | | | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |4,4µL | ||
| + | | | ||
| + | |- | ||
| + | |sum | ||
| + | |30µL | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | |||
| + | Ligation at 22,5°C for 30 min, denaturation at 65°C for 10 min. | ||
| + | |||
| + | Concentration of DNA in Ligation 1: | ||
| + | |||
| + | {| | ||
| + | |540ng | ||
| + | |A260/A280=2,4 | ||
| + | |} | ||
| + | |||
| + | |||
| + | <font color="#009933">Transformation</font> | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/18 Competent cells2|18 competent cells2]] | ||
| + | |||
| + | The incubation time for the cells is here 1 hour. | ||
| + | |||
== 9-09-2010 == | == 9-09-2010 == | ||
| - | + | ||
| + | |||
| + | <font color="#009933">Agarose gel electrophoresis of PCR 1, 3, 4a, 5, 7a, 9</font> | ||
| + | Agarose gel electrophoresis of PCR 1, 3, 4a, 5, 7a, 9 | ||
| + | |||
| + | -> protocol [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | results: "primer-clouds", PCR 9: no band | ||
| + | |||
| + | |||
| + | <font color="#009933">Plasmid extraction of ccdBcam, ccdBamp, Bak, CMV, PhiC31o</font> | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/4 Plasmid extraction from cells|4 Plasmid extraction from cells]] | ||
| + | |||
| + | {| | ||
| + | |Plasmid | ||
| + | |concentration [ng/µL] | ||
| + | |A260/A280 | ||
| + | |- | ||
| + | |ccdBcam | ||
| + | |22.5 | ||
| + | |1.0 | ||
| + | |- | ||
| + | |ccdBamp | ||
| + | |42.5 | ||
| + | |1.2 | ||
| + | |- | ||
| + | |Bak | ||
| + | |17.5 | ||
| + | |0.8 | ||
| + | |- | ||
| + | |CMV | ||
| + | |10.0 | ||
| + | |0.7 | ||
| + | |- | ||
| + | |PhiC310 | ||
| + | |60 | ||
| + | |1.4 | ||
| + | |} | ||
| + | |||
| + | Eluated with H<sub>2</sub>O instead of the Eluation Buffer. | ||
| + | |||
| + | |||
| + | |||
| + | <font color="#009933">New PCR 1, 3, 4a, 5, 7a, 9, 10 with phusion</font> | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! PCRnr. | ||
| + | ! 1 | ||
| + | ! 3 | ||
| + | ! 4a | ||
| + | ! 4b | ||
| + | ! 5 | ||
| + | ! 7a | ||
| + | ! 9 | ||
| + | ! 10 | ||
| + | |- | ||
| + | | template | ||
| + | | pDS7 (1:100); 4µl | ||
| + | | 2a; 0.9µl+2b; 0.6µl | ||
| + | | Bak; 0.5µl | ||
| + | | Bak; 0.5µl | ||
| + | | 4a; 0.8µl+4b; 1µl | ||
| + | | 190-6 (1:100); 1µl | ||
| + | | eGFP (1:25); 2µl | ||
| + | | PhiC31o (1:10); 1µl | ||
| + | |- | ||
| + | | primer [10pmol/µl] | ||
| + | | 1,2; 2.5µl | ||
| + | | 3,6; 2.5µl | ||
| + | | 7,8; 2.5µl | ||
| + | | 9,10; 2.5µl | ||
| + | | 7,10; 2.5µl | ||
| + | | 13,14; 2.5µl | ||
| + | | 20,21; 2.5µl | ||
| + | | 22,23; 2.5µl | ||
| + | |- | ||
| + | | H<sub>2</sub>O | ||
| + | | 28µl | ||
| + | | 30.5µl | ||
| + | | 31.5µl | ||
| + | | 31.5µl | ||
| + | | 30.2µl | ||
| + | | 31µl | ||
| + | | 30µl | ||
| + | | 31µl | ||
| + | |- | ||
| + | |Annealing temperature | ||
| + | |51.5°C | ||
| + | |53.1°C | ||
| + | |53.1°C | ||
| + | |51.5°C | ||
| + | |53.1°C | ||
| + | |57.5°C | ||
| + | |61°C | ||
| + | |57.5°C | ||
| + | |} | ||
| + | |||
| + | + in each assay: | ||
| + | |||
| + | dNTP mix: 1µl, 5x Phusion buffer: 10µl, DMSO:1.5µl, Phusion:0.5µl | ||
| + | |||
| + | sum: 50µl | ||
| + | |||
| + | Program: standard Phusion PCR, 29 cycles; annealing temperatures: see above | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/15 PCR with Phusion| 15 PCR with Phusion]] | ||
| + | |||
| + | [[Image: apo 09.09 1.jpg|400px|gel electrophoresis of PCR 1, 3, 4a, 5, 7a, 9, 10]] | ||
| + | |||
| + | [from left to right: PCR 1, 3, 4a, 5, 7a, 9, 10, Ladder] | ||
| + | |||
| + | <font color="#CC33CC">'''PCR3, 9, 10 with right bands.'''</font> | ||
| + | |||
| + | <font color="#009933">New PCR 1, 4a, 4b, 5, 7a with phusion</font> | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! PCRnr. | ||
| + | ! 1 | ||
| + | ! 4a | ||
| + | ! 4b | ||
| + | ! 5 | ||
| + | ! 7a | ||
| + | |- | ||
| + | | template | ||
| + | | pDS7 (1:10); 1µl | ||
| + | | Bak 1 ; 1µl | ||
| + | | Bak 1; 1µl | ||
| + | | 4a, 4b; 2 x 1.5µl | ||
| + | | pCT 190-6 (1:40); 1µl | ||
| + | |- | ||
| + | | primer | ||
| + | | 1, 2; 2 x 2.5µl | ||
| + | | 7, 8; 2 x 2.5µl | ||
| + | | 9, 10; 2 x 2.5µl | ||
| + | | 7, 10; 2 x 2.5µl | ||
| + | | 13, 14; 2x 2.5µl | ||
| + | |- | ||
| + | | H<sub>2</sub>O | ||
| + | | 31µl | ||
| + | | 31µl | ||
| + | | 31µl | ||
| + | | 30µl | ||
| + | | 30µl | ||
| + | |- | ||
| + | | Annealing temperature | ||
| + | | 52°C | ||
| + | | 52°C | ||
| + | | 48°C | ||
| + | | 50°C | ||
| + | | 55°C | ||
| + | |} | ||
| + | |||
| + | + each assay with dNTP-mix (1µl), 5xBuffer (10µl, DMSO (1.5µl), Phusion (0.5µl) | ||
| + | -> sum: 50µl | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/17 Special thermal cycler programs/Touch down phusion|Touch down 59 with phusion]], 30 cycles with gradient appropriate for the annealing temperatures above. | ||
| + | |||
== 9-10-2010 == | == 9-10-2010 == | ||
| - | + | <font color="#009933">Agarose gel electrophoresis of PCR 1, 4a, 4b, 5, 7a</font> | |
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | [[Image: 9-10-10.jpg|400px|gel electrophoresis of PCR 1, 4a, 4b, 5, 7a]] | ||
| + | |||
| + | [From left to right: Ladder, 1, 4a, 4b, 5, 7a] | ||
| + | |||
| + | Only<font color="#CC33CC"> '''4a has been amplified successfully.'''</font> | ||
| + | |||
| + | |||
| + | <font color="#009933">Agarose gel extraction of PCR 3, 4a, 5, 9, 10</font> | ||
| + | |||
| + | [[Image: apo-10-9-10-2.jpg|400px|gel electrophoresis of PCR 3, 4a, 5, 9, 10]] | ||
| + | |||
| + | from left to right:<font color="#CC33CC"> '''PCR 3, band ~700bp, PCR 4a, band ~300bp, PCR 5, bands ~550bp (5*), ~650bp (5), PCR 9, band ~800bp, PCR 10, band ~1900bp'''</font> | ||
| + | |||
| + | results: | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! | ||
| + | ! PCR 3 | ||
| + | ! PCR 4a | ||
| + | ! PCR 5 | ||
| + | ! PCR 5* | ||
| + | ! PCR 9 | ||
| + | ! PCR 10 | ||
| + | |- | ||
| + | | concentration [ng/µl] | ||
| + | | 20 | ||
| + | | 10 | ||
| + | | 5 | ||
| + | | 30 | ||
| + | | 35 | ||
| + | | 25 | ||
| + | |- | ||
| + | | A260/A280 | ||
| + | | 1.6 | ||
| + | | 1.333 | ||
| + | | 2.0 | ||
| + | | 2.0 | ||
| + | | 1.750 | ||
| + | | 2.0 | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/12 Gel extraction or PCR Clean up| 12 Gel extraction or PCR Clean up (Promega kit)]] | ||
| + | |||
== 9-11-2010 == | == 9-11-2010 == | ||
weekend | weekend | ||
| Line 642: | Line 2,570: | ||
== 9-13-2010 == | == 9-13-2010 == | ||
| - | + | charges for sequencing (retry of 6.9.) | |
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! name | ||
| + | ! 4a-7 | ||
| + | ! 4a-8 | ||
| + | ! 4b-9 | ||
| + | ! 4b-10 | ||
| + | ! 3-3 | ||
| + | ! 3-6 | ||
| + | ! 6-11 | ||
| + | ! 6-12 | ||
| + | |- | ||
| + | | DNA | ||
| + | | 4a; 2.4µl | ||
| + | | 4a; 2.4µl | ||
| + | | 4b; 0.5µl | ||
| + | | 4b; 0.5µl | ||
| + | | 3; 2µl | ||
| + | | 3; 2µl | ||
| + | | 6; 0.5µl | ||
| + | | 6; 0.5µl | ||
| + | |- | ||
| + | | primer [1pmol/µl] | ||
| + | | 7; 3.2µl | ||
| + | | 8; 3.2µl | ||
| + | | 9; 3.2µl | ||
| + | | 10; 3.2µl | ||
| + | | 3; 3.2µl | ||
| + | | 6; 3.2µl | ||
| + | | 11; 3.2µl | ||
| + | | 12; 3.2µl | ||
| + | |- | ||
| + | | Tris (10mM); pH 8.2 | ||
| + | | 1.4µl | ||
| + | | 1.4µl | ||
| + | | 3.3µl | ||
| + | | 3.3µl | ||
| + | | 1.8µl | ||
| + | | 1.8µl | ||
| + | | 3.3µl | ||
| + | | 3.3µl | ||
| + | |} | ||
| + | every charge 7µl | ||
| + | |||
| + | |||
| + | <font color="#009933">New PCR7a with Taq</font> | ||
| + | |||
| + | template: p190-6 | ||
| + | |||
| + | Primer 13,14 | ||
| + | |||
| + | Mixture | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |template (~4ng) | ||
| + | |2µl (1:100) | ||
| + | |- | ||
| + | |MasterMix for Taq | ||
| + | |10µl | ||
| + | |- | ||
| + | |Primer *2 | ||
| + | |1.5µl *2 | ||
| + | |- | ||
| + | |DMSO | ||
| + | |0,5µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |4.5µl | ||
| + | |- | ||
| + | |sum | ||
| + | |20µl | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | PCR program: touchdown PCR with Taq | ||
| + | |||
| + | ::{| | ||
| + | |1: 94°C 2' | ||
| + | ---- | ||
| + | |- | ||
| + | |2: 94°C 30" | ||
| + | |- | ||
| + | |3: 64°C/62°C/60°C/58°C/56°C/54° 30" | ||
| + | |- | ||
| + | |4: 72°C 2' | ||
| + | |- | ||
| + | |5: (for each temperature)repeat 2-4 2x | ||
| + | ---- | ||
| + | |- | ||
| + | |6: 94°C 30" | ||
| + | |- | ||
| + | |7: 58°C 30" | ||
| + | |- | ||
| + | |8: 72°C 2' | ||
| + | |- | ||
| + | |9: repeat 6-8 29x | ||
| + | ---- | ||
| + | |- | ||
| + | |10: 72°C 10' | ||
| + | |- | ||
| + | |11: 15°C break | ||
| + | |} | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/21 PCR with Taq Mastermix|21 PCR with Taq Mastermix]] | ||
| + | |||
| + | <font color="#009933">overnight culture inoculated of</font> | ||
| + | |||
| + | :CMV (amp) | ||
| + | :ccdB (amp) | ||
| + | :ccdB (cam) | ||
| + | :pC31o (amp) | ||
| + | :Bak (amp) | ||
| + | |||
| + | |||
| + | <font color="#009933">New PCR7a with Phusion Hot Start</font> | ||
| + | |||
| + | with 2ng and 4 ng template | ||
| + | |||
| + | ::{| | ||
| + | |- | ||
| + | |template | ||
| + | |2ng | ||
| + | |4ng | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |54µl | ||
| + | |53µl | ||
| + | |- | ||
| + | |Buffer 5x | ||
| + | |20µl | ||
| + | |20µl | ||
| + | |- | ||
| + | |Primer (13,14) | ||
| + | |2*10µl | ||
| + | |2*10µl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |3µl | ||
| + | |3µl | ||
| + | |- | ||
| + | |Hot Start | ||
| + | |1µl | ||
| + | |1µl | ||
| + | |} | ||
| + | |||
| + | total: 100µl each, divided into 5 charges | ||
| + | |||
| + | program: | ||
| + | |||
| + | ::98°C 30sec | ||
| + | ::---- | ||
| + | ::98°C 10sec | ||
| + | ::gradient: 50°C, 53°C, 56,6°C, 60,2°C, 64,5°C 30sec 30 cycles | ||
| + | ::72 15sec | ||
| + | ::---- | ||
| + | ::72°C 5min | ||
| + | ::12°C forever | ||
| + | |||
| + | -> Protocol:[[Team:LMU-Munich/Notebook/Protocols/20 PCR with Phusion Hot Start|20 PCR with Phusion Hot Start]] | ||
| + | |||
== 9-14-2010 == | == 9-14-2010 == | ||
| - | + | <font color="#009933">Agarose gelelectrophoresis of PCR 7a</font> | |
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | bands 1 to 5: 2ng of template DNA | ||
| + | |||
| + | bands 6 to 10: 4ng of template DNA | ||
| + | |||
| + | -> no bands | ||
| + | |||
| + | <font color="#009933">new PCR 7a and PCR 8 (without mutation)</font> | ||
| + | with mastermix, without DMSO | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |number: | ||
| + | |7a diluted | ||
| + | |7a undiluted | ||
| + | |8 | ||
| + | |- | ||
| + | |mastermix | ||
| + | |50µl | ||
| + | |50µl | ||
| + | |10µl | ||
| + | |- | ||
| + | |primer | ||
| + | |2*10µl | ||
| + | |2*10µl | ||
| + | |2*2µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |29µl | ||
| + | |29µl | ||
| + | |5µl | ||
| + | |- | ||
| + | |template | ||
| + | |1µl 190-6 (1:100) | ||
| + | |1µl 190-6 | ||
| + | |1µl 190-6 (1:100) | ||
| + | |} | ||
| + | |||
| + | 7a diluated and undiluated: divided into 3 charges | ||
| + | |||
| + | 7a diluated: 1,2,3; 7a undiluated: 4,5,6 | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |1 | ||
| + | |2 | ||
| + | |3 | ||
| + | |4 | ||
| + | |5 | ||
| + | |6 | ||
| + | |8 | ||
| + | |- | ||
| + | |48°C | ||
| + | |52°C | ||
| + | |56,1°C | ||
| + | |48°C | ||
| + | |52°C | ||
| + | |56,1°C | ||
| + | |52°C | ||
| + | |} | ||
| + | |||
| + | program: | ||
| + | |||
| + | ::{| | ||
| + | |94°C 2' | ||
| + | ---- | ||
| + | |- | ||
| + | |94°C 30" | ||
| + | |- | ||
| + | |48°C/52°C/56,1°C 30" | ||
| + | |- | ||
| + | |72°C 1,5' | ||
| + | |- | ||
| + | |(for each temperature)repeat 30x | ||
| + | ---- | ||
| + | |- | ||
| + | |72°C 5' | ||
| + | |} | ||
| + | |||
| + | <font color="#009933">Plasmid extraction of ccdB (amp) and ccdB (cam)</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/4 Plasmid extraction from cells|4 Plasmid extraction from cells]] | ||
| + | |||
| + | results: | ||
| + | |||
| + | ::ccdB(amp): 105ng/µl A260/280: 2,00 | ||
| + | ::cddB(cam): 102ng/µl A260/280: 1,952 | ||
| + | |||
== 9-15-2010 == | == 9-15-2010 == | ||
| - | + | <font color="#009933">Agarose gelelectrophoresis of PCR 7a diluted &7a undiluted & "8"(without mutation)</font> | |
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | [[Image:9 15 2010.jpg|400px|9 15 10 Gelphoto]] | ||
| + | |||
| + | from left to right: 7a: 1:100 diluted: 48°C, 52°C, 56.1°C, undiluted: 48°C, 52°C, 56.1°C, 8 without Mutation | ||
| + | |||
| + | <font color="#CC33CC">'''weak right band (with fuzz)for 7a '''</font>undiluated with best result for 56°C | ||
| + | |||
| + | -> <font color="#009933">new PCR 7a and "8"</font> | ||
| + | |||
| + | with mastermix Taq | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/21 PCR with Taq Mastermix|21 PCR with Taq Mastermix]] | ||
| + | |||
| + | template: 190-6, 1:10 and 1:5 | ||
| + | |||
| + | temp: 56°C and 58°C | ||
| + | |||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |number | ||
| + | |7a | ||
| + | |7a | ||
| + | |"8" | ||
| + | |"8" | ||
| + | |- | ||
| + | |Mastermix | ||
| + | |20µl | ||
| + | |20µl | ||
| + | |20µl | ||
| + | |20µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |15µl | ||
| + | |15µl | ||
| + | |15µl | ||
| + | |15µl | ||
| + | |- | ||
| + | |Primer | ||
| + | |(13,14) 2*2µl | ||
| + | |(13,14) 2*2µl | ||
| + | |(13,16) 2*2µl | ||
| + | |(13,16) 2*2µl | ||
| + | |- | ||
| + | |Template | ||
| + | |190-6 (1:5) 1µl | ||
| + | |190-6 (1:10) 1µl | ||
| + | |190-6 (1:5) 1µl | ||
| + | |190-6 (1:10) 1µl | ||
| + | |} | ||
| + | |||
| + | 2 charges each: one for 56°C and one for 58°C annealing temperature | ||
| + | |||
| + | |||
| + | program: | ||
| + | |||
| + | ::{| | ||
| + | |94°C 2' | ||
| + | ---- | ||
| + | |- | ||
| + | |94°C 30" | ||
| + | |- | ||
| + | |56°C/58°C 30" | ||
| + | |- | ||
| + | |72°C 1,5' | ||
| + | |- | ||
| + | |(for each temperature)repeat 30x | ||
| + | ---- | ||
| + | |- | ||
| + | |72°C 5' | ||
| + | |} | ||
| + | |||
| + | |||
| + | <font color="#009933">Restriction Digestion of PCR 1 (17.8.), PCR 3 (10.9.), PCR 5<sub>1</sub> (10.9.), PCR 5<sub>2</sub> (10.9.), PCR 6 (17.8.), PCR 9 (10.9.), PCR 10 (30.8.) for ligation with vector</font> | ||
| + | |||
| + | EcoR1 + Pst1 with Buffer H; 50ng DNA | ||
| + | |||
| + | {| | ||
| + | | | ||
| + | |1 | ||
| + | |3 | ||
| + | |5<sub>1</sub> | ||
| + | |5<sub>2</sub> | ||
| + | |6 | ||
| + | |9 | ||
| + | |10 | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |13µl | ||
| + | |12,5µl | ||
| + | |5µl | ||
| + | |13µl | ||
| + | |13,5µl | ||
| + | |13,5µl | ||
| + | |12,5µl | ||
| + | |- | ||
| + | |Buffer H | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |- | ||
| + | |BSA (1:10) | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |- | ||
| + | |DNA | ||
| + | |2µl | ||
| + | |2,5µl | ||
| + | |10µl | ||
| + | |2µl | ||
| + | |1,5µl | ||
| + | |1,5µl | ||
| + | |2,5µl | ||
| + | |- | ||
| + | |EcoR1 | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |- | ||
| + | |Pst1 | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |} | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/5 Restriction digest|5 Restriction digestion]] | ||
| + | |||
| + | <font color="#009933">Agarose gelelectrophoresis of PCR 7a 1-4 & "8" 1-4(without mutation)</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | -> bad results | ||
| + | |||
| + | <font color="#009933">purification of restriction digestion</font> | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | | | ||
| + | |concentration (ng/µl) | ||
| + | |A<sub>260</sub>/A<sub>280</sub> | ||
| + | |(supposed) lenght | ||
| + | |- | ||
| + | |1 | ||
| + | |20 | ||
| + | |1,000 | ||
| + | |492 | ||
| + | |- | ||
| + | |3 | ||
| + | |5 | ||
| + | |1,000 | ||
| + | |1087 | ||
| + | |- | ||
| + | |5<sub>1</sub> | ||
| + | |22,5 | ||
| + | |1,8 | ||
| + | |688 | ||
| + | |- | ||
| + | |5<sub>2</sub> | ||
| + | |7,5 | ||
| + | |1,5 | ||
| + | |688 | ||
| + | |- | ||
| + | |6 | ||
| + | |10 | ||
| + | |1,333 | ||
| + | |237 | ||
| + | |- | ||
| + | |9 | ||
| + | |20 | ||
| + | |1,6 | ||
| + | |808 | ||
| + | |- | ||
| + | |10 | ||
| + | |17,5 | ||
| + | |1,75 | ||
| + | |1888 | ||
| + | |} | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/12 Gel extraction or PCR Clean up| 12 Gel extraction or PCR Clean up (Promega kit)]] | ||
| + | |||
| + | <font color="#009933">Ligation with pSB1C3 (2072bp)</font> | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |for | ||
| + | |# | ||
| + | |backbone | ||
| + | |insert (µl) | ||
| + | |charge (µl) | ||
| + | |Buffer 10x (µl) | ||
| + | |H<sub>2</sub>O (µl) | ||
| + | |- | ||
| + | |100ng | ||
| + | |1 | ||
| + | |4 | ||
| + | |7,12 | ||
| + | |20 | ||
| + | |2 | ||
| + | |6,38 | ||
| + | |- | ||
| + | |50ng | ||
| + | |3 | ||
| + | |2 | ||
| + | |31,48 | ||
| + | |40 | ||
| + | |4 | ||
| + | |2,02 | ||
| + | |- | ||
| + | |100ng | ||
| + | |5<sub>1</sub> | ||
| + | |4 | ||
| + | |8,85 | ||
| + | |20 | ||
| + | |2 | ||
| + | |4,65 | ||
| + | |- | ||
| + | |100ng | ||
| + | |5<sub>2</sub> | ||
| + | |4 | ||
| + | |26,56 | ||
| + | |40 | ||
| + | |4 | ||
| + | |4,94 | ||
| + | |- | ||
| + | |100ng | ||
| + | |6 | ||
| + | |4 | ||
| + | |6,86 | ||
| + | |20 | ||
| + | |2 | ||
| + | |13,36 | ||
| + | |- | ||
| + | |100ng | ||
| + | |9 | ||
| + | |4 | ||
| + | |11,7 | ||
| + | |20 | ||
| + | |2 | ||
| + | |1,8 | ||
| + | |- | ||
| + | |100ng | ||
| + | |10 | ||
| + | |4 | ||
| + | |31,24 | ||
| + | |40 | ||
| + | |4 | ||
| + | |4,26 | ||
| + | |} | ||
| + | plus 1 µl T4 ligase in each charge | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/22 Ligation|22 Ligation]] | ||
| + | |||
| + | |||
| + | <font color="#009933">new PCR 7a & "8"</font> | ||
| + | |||
| + | with mastermix Taq 56°C | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | | | ||
| + | |7a-1 | ||
| + | |7a-2 | ||
| + | |"8"-1 | ||
| + | |"8"-2 | ||
| + | |- | ||
| + | |mastermix (µl) | ||
| + | |10 | ||
| + | |10 | ||
| + | |10 | ||
| + | |10 | ||
| + | |- | ||
| + | |template | ||
| + | |190-6 1µl | ||
| + | |190-6 2µl | ||
| + | |190-6(1:10) 1µl | ||
| + | |190-6(1:10) 2µl | ||
| + | |- | ||
| + | |primer | ||
| + | |13&14 2*1µl | ||
| + | |13&14 2*1µl | ||
| + | |13&16 2*1µl | ||
| + | |13&16 2*1µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |7µl | ||
| + | |6µl | ||
| + | |7µl | ||
| + | |6µl | ||
| + | |} | ||
| + | sum: 20µl each | ||
| + | |||
| + | pcr-program: | ||
| + | ::{| | ||
| + | |94°C 2' | ||
| + | ---- | ||
| + | |- | ||
| + | |94°C 30" | ||
| + | |- | ||
| + | |56°C 30" | ||
| + | |- | ||
| + | |72°C 1,5' | ||
| + | |- | ||
| + | |(for each temperature)repeat 30x | ||
| + | ---- | ||
| + | |- | ||
| + | |72°C 5' | ||
| + | |- | ||
| + | |12°C forever | ||
| + | |} | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/21 PCR with Taq Mastermix|21 PCR with Taq Mastermix]] | ||
| + | |||
== 9-16-2010 == | == 9-16-2010 == | ||
| - | + | <font color="#009933">Agarose gelelectrophoresis of PCR 7a-1, PCR 7a-2, PCR 8-1, 8-2 and of all three PCR 6 we ever made (in order to control whether we have extracted the right fragment (237) or just the primerdimers)</font> | |
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | [[Image:9 16 10.jpg|400px|9 16 10 Gelphoto]] | ||
| + | |||
| + | From left to right: 6<sub>2</sub>, 6<sub>1</sub>, 6, Ladder, 7a-1, 7a-2, 8-2, 8-1 | ||
| + | |||
| + | ::results: | ||
| + | |||
| + | ::6-1 and 6-2: ? | ||
| + | |||
| + | ::<font color="#CC33CC">'''6: ok'''</font> | ||
| + | ::7a-1,7a-2,"8"-1,"8"-2: primerdimer-problem -> we ordered new primers! | ||
| + | |||
| + | <font color="#009933">transformation of the ligations 1,3,5<sub>1</sub>, 5<sub>2</sub>, 6, 9, 10</font> | ||
| + | |||
| + | -> Protocol:[[Team:LMU-Munich/Notebook/Protocols/3 Transformation|3 Transformation]] | ||
| + | |||
| + | ->plated on plates with chloramphenicol | ||
| + | |||
| + | <font color="#009933">again PCR 1,3,5,6,9,10 in order to increase the profit (Ausbeute) for ligation</font> | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | | | ||
| + | |1 | ||
| + | |3 | ||
| + | |5 | ||
| + | |6 | ||
| + | |9 | ||
| + | |10 | ||
| + | |- | ||
| + | |template | ||
| + | |prev TRE(1:30) 1µl | ||
| + | |2a(1:5) 1µl + 2b 0,5µl | ||
| + | |4a 0,5µl + 4b 0,5µl | ||
| + | |SV40PA (pcDNA3)(1:50) 1µl | ||
| + | |eGFP(1:5) 1µl | ||
| + | |PhiC310(1:6) 1µl | ||
| + | |- | ||
| + | |primer | ||
| + | |1&2 2*2,5µl | ||
| + | |3&6 2*2,5µl | ||
| + | |7&10 2*2,5µl | ||
| + | |11&12 2*2,5µl | ||
| + | |20&21 2*2,5µl | ||
| + | |22&23 2*2,5µl | ||
| + | |- | ||
| + | |T<sub>m</sub> | ||
| + | |54°C | ||
| + | |50°C | ||
| + | |49°C | ||
| + | |56°C | ||
| + | |58°C | ||
| + | |58°C | ||
| + | |- | ||
| + | |mastermix (µl) | ||
| + | |25 | ||
| + | |25 | ||
| + | |25 | ||
| + | |25 | ||
| + | |25 | ||
| + | |25 | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,25µl | ||
| + | |1,25µl | ||
| + | |1,25µl | ||
| + | |1,25µl | ||
| + | |1,25µl | ||
| + | |1,25µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O (µl) | ||
| + | |17,75 | ||
| + | |17,25 | ||
| + | |17,75 | ||
| + | |17,75 | ||
| + | |17,75 | ||
| + | |17,75 | ||
| + | |} | ||
| + | |||
| + | pcr-program: | ||
| + | |||
| + | ::{| | ||
| + | |94°C 2' | ||
| + | ---- | ||
| + | |- | ||
| + | |94°C 30" | ||
| + | |- | ||
| + | |T<sub>m</sub> (see above) 30" | ||
| + | |- | ||
| + | |72°C 1,5' | ||
| + | |- | ||
| + | |(for each temperature)repeat 30x | ||
| + | ---- | ||
| + | |- | ||
| + | |72°C 5' | ||
| + | |} | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/21 PCR with Taq Mastermix|21 PCR with Taq Mastermix]] | ||
| + | |||
== 9-17-2010 == | == 9-17-2010 == | ||
| - | + | <font color="#009933">Analysis of the transformation from yesterday</font> | |
| + | |||
| + | Colonies on plates when 100µl or pellet plated: | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | | | ||
| + | |1 | ||
| + | |3 | ||
| + | |5 | ||
| + | |6 | ||
| + | |9 | ||
| + | |10 | ||
| + | |- | ||
| + | |pellet | ||
| + | |no | ||
| + | |yes | ||
| + | |yes | ||
| + | |yes | ||
| + | |yes | ||
| + | |yes | ||
| + | |yes | ||
| + | |- | ||
| + | |100µl | ||
| + | |no | ||
| + | |yes | ||
| + | |yes | ||
| + | |yes | ||
| + | |yes | ||
| + | |yes | ||
| + | |yes | ||
| + | |} | ||
| + | |||
| + | <font color="#009933">again: transformation of ligation 1</font> | ||
| + | |||
| + | ->Protocol: [[Team:LMU-Munich/Notebook/Protocols/3 Transformation|3 Transformation]] | ||
| + | |||
| + | <font color="#009933">Agarose gelelectrophoresis of yesterday's PCR 1,3,5,6,9,10(without mutation)</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | [[Image:9 17 10apo.jpg| 400px |9 17 10 Gelphoto]] | ||
| + | |||
| + | From left to right: PCR1, PCR3, PCR5, Ladder, PCR6, PCR9, PCR10 | ||
| + | |||
| + | -> <font color="#CC33CC">'''right band for PCR 3,5,6,9,10'''</font>; no band for PCR 1 | ||
| + | |||
| + | [[Image:9 17 10apo2.jpg| 400px |9 17 10 Gelphoto2]] | ||
| + | |||
| + | From left to right: Ladder, PCR3, PCR5, PCR9, PCR10 | ||
| + | |||
| + | Bands took: 3<sub>2</sub>: Band short over 1000bp, 3<sub>1</sub>: Band short under 1000bp (probably 3<sub>2</sub> right), 5 the highest band, 9 upper band, 10 upper band | ||
| + | |||
| + | <font color="#009933">new PCR 1,5,6</font> | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | | | ||
| + | |1 | ||
| + | |5 | ||
| + | |6 | ||
| + | |- | ||
| + | |template | ||
| + | |pTRE Rev 0,5µl | ||
| + | |4a 1,5µl + 4b 1,5µl | ||
| + | |pcDNA3 0,5µl | ||
| + | |- | ||
| + | |primer | ||
| + | |1&2 2*2,5µl | ||
| + | |7&10 2*2,5µl | ||
| + | |11&12 2*2,5µl | ||
| + | |- | ||
| + | |buffer 10x | ||
| + | |5µl | ||
| + | |5µl | ||
| + | |5µl | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |1µl | ||
| + | |1µl | ||
| + | |1µl | ||
| + | |- | ||
| + | |Pfu | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |0,5µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |26,75 µl | ||
| + | |24,25 µl | ||
| + | |26,75 µl | ||
| + | |- | ||
| + | |T<sub<m</sub> | ||
| + | |51°C | ||
| + | |50°C | ||
| + | |55°C | ||
| + | |} | ||
| + | |||
| + | -> Protocol: | ||
| + | pcr-program: standard Pfu [[Team:LMU-Munich/Notebook/Protocols/10 PCR with Pfu| 10 PCR with Pfu]] | ||
| + | |||
| + | |||
| + | <font color="#009933">Restriction digestion of PCR products, ccdB Plasmids and Biobricks for the 3A Method</font> | ||
| + | |||
| + | |||
| + | {| | ||
| + | |Name | ||
| + | |H<sub>2</sub>O | ||
| + | |Buffer (each 2µl) | ||
| + | |BSA (1:10) | ||
| + | |DNA Volume | ||
| + | |DNA Mass | ||
| + | |Enzyms (2*0.5µl) | ||
| + | |- | ||
| + | |ccdBa | ||
| + | |25 µl | ||
| + | |H | ||
| + | |2µl | ||
| + | |20µl | ||
| + | |~600ng | ||
| + | |E+P | ||
| + | |- | ||
| + | |ccdBa | ||
| + | |25µl | ||
| + | |H | ||
| + | |2µl | ||
| + | |20µl | ||
| + | |~700ng | ||
| + | |E+P | ||
| + | |- | ||
| + | |1 | ||
| + | |43µl | ||
| + | |MC | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |~500ng | ||
| + | |E+S | ||
| + | |- | ||
| + | |3 | ||
| + | |25µl | ||
| + | |B | ||
| + | |2µl | ||
| + | |20µl | ||
| + | |~200ng | ||
| + | |X+P | ||
| + | |- | ||
| + | |Primer 18+19 | ||
| + | |25µl | ||
| + | |B | ||
| + | |2µl | ||
| + | |10µl+10µl | ||
| + | |? | ||
| + | |X+P | ||
| + | |- | ||
| + | |5<sub>1</sub> | ||
| + | |25µl | ||
| + | |MC | ||
| + | |2µl | ||
| + | |20µl | ||
| + | |~100ng | ||
| + | |E+S | ||
| + | |- | ||
| + | |5<sub>2</sub> | ||
| + | |25µl | ||
| + | |MC | ||
| + | |2µl | ||
| + | |20µl | ||
| + | |~600ng | ||
| + | |E+S | ||
| + | |- | ||
| + | |6 | ||
| + | |35µl | ||
| + | |B | ||
| + | |2µl | ||
| + | |10µl | ||
| + | |~400ng | ||
| + | |X+P | ||
| + | |- | ||
| + | |9 | ||
| + | |25µl | ||
| + | |MC | ||
| + | |2µl | ||
| + | |20µl | ||
| + | |~700ng | ||
| + | |E+S | ||
| + | |- | ||
| + | |10 | ||
| + | |25µl | ||
| + | |MC | ||
| + | |2µl | ||
| + | |20µl | ||
| + | |~400ng | ||
| + | |E+S | ||
| + | |- | ||
| + | |eGFP | ||
| + | |35µl | ||
| + | |MC | ||
| + | |2µl | ||
| + | |10µl | ||
| + | |~550ng | ||
| + | |E+S | ||
| + | |- | ||
| + | |CMV1 | ||
| + | |35µl | ||
| + | |H | ||
| + | |2µl | ||
| + | |10µl | ||
| + | |~530ng | ||
| + | |E+P | ||
| + | |} | ||
| + | |||
| + | -> Protocol:[[Team:LMU-Munich/Notebook/Protocols/5 Restriction digest|5 Restriction digestion]] | ||
| + | |||
| + | <font color="#009933">Colony PCR of Ligations 3, 5<sub>1</sub>, 5<sub>2</sub>, 6, 9, 10 and PCR of CMV1 (to test if right)</font> | ||
| + | |||
| + | 10 1-4, 9 1-4, 6 1-4, 5<sub>1</sub> 1-4, 5<sub>2</sub> 1-4, 3 1-4 Colonies picked and put in following Mix: | ||
| + | |||
| + | {| | ||
| + | |PCR Mastermix | ||
| + | |10µl | ||
| + | |- | ||
| + | |Biobrick Primer F | ||
| + | |1.5µl | ||
| + | |- | ||
| + | |Biobrick Primer R | ||
| + | |1.5µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |7µl | ||
| + | |} | ||
| + | |||
| + | CMV1: | ||
| + | |||
| + | {| | ||
| + | |PCR Mastermix | ||
| + | |10µl | ||
| + | |- | ||
| + | |Biobrick Primer F | ||
| + | |1.5µl | ||
| + | |- | ||
| + | |Biobrick Primer R | ||
| + | |1.5µl | ||
| + | |- | ||
| + | |Template | ||
| + | |2µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |5µl | ||
| + | |} | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/21 PCR with Taq Mastermix|21 PCR with Taq Mastermix]] | ||
| + | |||
| + | <font color="#009933">Agarose Gel electrophoresis of 3, 5<sub>1</sub>, 5<sub>2</sub>, 6, 9, 10 and CMV1 (to test if right)</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | [[Image:17 9 10apo.jpg|400px|17 9 10 Gelphoto]] | ||
| + | |||
| + | Above: From left to right: 3.1, 3.2, 3.3, 3.4, 5<sub>1</sub>.1, 5<sub>1</sub>.2, 5<sub>1</sub>.3, 5<sub>1</sub>.4, Ladder, 5<sub>2</sub>.1, 5<sub>2</sub>.2, 5<sub>2</sub>.3, 5<sub>2</sub>.4 | ||
| + | |||
| + | Below: From left to right: 6.1, 6.2, 6.3, 6.4, 9.1, 9.2, 9.3, 9.4, Ladder, 10.1, 10.2, 10.3, 10.4, CMV1 | ||
| + | |||
| + | -> <font color="#CC33CC">'''right bands for 6.3 and 6.4, CMV1 ~1200bp (we think that this is right, as Biobrick sequenzing information indicates that it isn't 654bp but about 1200bp)'''</font> | ||
| + | |||
| + | |||
| + | <font color="#009933">PCR clean up of PCR Gel extraction 3<sub>1</sub>, 3<sub>2</sub>, 5, 9, 10, PCR Product 6 and digestion PCR1, Primer 18+19, R5<sub>1</sub>, R5<sub>2</sub>, PCR6, PCR9, PCR10, PCR3</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/12 Gel extraction or PCR Clean up| 12 Gel extraction or PCR Clean up (Promega kit)]] | ||
| + | |||
== 9-18-2010 == | == 9-18-2010 == | ||
weekend | weekend | ||
| Line 657: | Line 3,536: | ||
== 9-20-2010 == | == 9-20-2010 == | ||
| - | + | <font color="#009933">Dephosphorylation of linearized ccdB amp and ccdB cam</font> | |
| + | |||
| + | Add 1µl TSAP to digested vectors. | ||
| + | |||
| + | Incutation: 37°C 15min; Inhibition: 74°C 15min | ||
| + | |||
| + | <font color="#009933">Ligation and 3A-Assemblies</font> | ||
| + | |||
| + | Jump-or-Die Ligations: 1a, 1b (5<sub>1</sub>),1b (5<sub>2</sub>), 2, 3 | ||
| + | |||
| + | Cut'N'Survive Ligations: 1a, 2b, | ||
| + | |||
| + | Biobricks for both Systems: CMV, PCR1 (tet-on-promoter) | ||
| + | |||
| + | Mixtures: | ||
| + | |||
| + | - 3A Assemblies (everything except CMV, PCR1): | ||
| + | |||
| + | Inserts: each 8µl; ccdB vector: 1µl; T4 Ligase: 0.5µl; T4 Ligase Buffer: 2µl; H<sub>2</sub>O: 1µl | ||
| + | |||
| + | - Biobricks: | ||
| + | |||
| + | Insert: 10µl; ccdB vector: 1µl; T4 Ligase: 0.5µl; T4 Ligase Buffer: 2µl; H<sub>2</sub>O: 7µl | ||
| + | |||
| + | Incubation: 2:30h 16°C; Inhibition: 10 min 65°C | ||
| + | |||
| + | ->Protocols: [[Team:LMU-Munich/Notebook/Protocols/22 Ligation|22 Ligation]], [[Team:LMU-Munich/Notebook/Protocols/13 3A Method for Biobrick assembly| 13 3A Method for Biobrick assembly]] | ||
| + | |||
| + | <font color="#009933">Colony PCR of PCR product ligations with pSB1C3</font> | ||
| + | |||
| + | Mixture: | ||
| + | |||
| + | PCR Mastermix:130µl ; PrimerF:19,5µl ; PrimerR:19,5µl ; H<sub>2</sub>O:91µl | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/21 PCR with Taq Mastermix|21 PCR with Taq Mastermix]] | ||
| + | |||
| + | <font color="#009933">Agarose Gel Electrophoresis of Colony PCRs</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | [[Image:20.9.10.jpg|400px|Colony PCRs]] | ||
| + | |||
| + | above from left to right: pathway/ladder/3.5/3.6/3.7/3.8/5<sub>1</sub>.5/5<sub>1</sub>.6/5<sub>1</sub>.7/5<sub>1</sub>.8/5<sub>2</sub>.5/5<sub>2</sub>.6/5<sub>2</sub>.7/5<sub>2</sub>.8 | ||
| + | |||
| + | below from left to right: ladder/6.5/6.6/6.7/6.8/9.5/9.6/9.7/9.8/10.5/10.6/10.7/10.8 | ||
| + | |||
== 9-21-2010 == | == 9-21-2010 == | ||
| - | + | <font color="#009933">Agarose Gel Electrophoresis of PCRs 1, 5, 6, and Colony PCR 3.8, 5<sub>1</sub>.7, 5<sub>2</sub>.6, 9.7, 10.5, 10.6</font> | |
| + | |||
| + | -> Bad results, probably too much cells in PCR mix | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | <font color="#009933">Colony PCR of 3.8, 5<sub>1</sub>.7, 5<sub>2</sub>.6, 9.7, 10.5, 10.6 again and of the 3A Ligations of yesterday as well as the Ligation of PCR1 and CMV with pSB1C3</font> | ||
| + | |||
| + | PCR Master Mix: 60µl | ||
| + | |||
| + | Primer F: 9µl | ||
| + | |||
| + | Primer R: 9µl | ||
| + | |||
| + | H<sub>2</sub>O: 42µl | ||
| + | |||
| + | ->22charges à 5µl | ||
| + | |||
| + | -> Protocol:[[Team:LMU-Munich/Notebook/Protocols/21 PCR with Taq Mastermix|21 PCR with Taq Mastermix]] | ||
| + | |||
| + | |||
| + | <font color="#009933">PCR 7a with new Primers</font> | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |template 190-6 (1:10) | ||
| + | |1µl | ||
| + | |- | ||
| + | |Master Mix | ||
| + | |25µl | ||
| + | |- | ||
| + | |Primer 13k (short) | ||
| + | |3,75µl | ||
| + | |- | ||
| + | |Primer 14 | ||
| + | |3,75µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |16,5µl | ||
| + | |} | ||
| + | |||
| + | sum: 50µl | ||
| + | |||
| + | same with Primer 13l (long) | ||
| + | |||
| + | ->Protocol: [[Team:LMU-Munich/Notebook/Protocols/21 PCR with Taq Mastermix|21 PCR with Taq Mastermix]] | ||
| + | |||
| + | <font color="#009933">Agarose Gel Electrophoresis of Colony PCRs and PCR 7a</font> | ||
| + | different ladder: [[image:fermentas ladder mix.jpg|thumb|right|fermentas dna ladder mix (source:http://www.fermentas.com/en/products/all/dna-electrophoresis/generuler-dna-ladders/sm0333-generuler-mix]] | ||
| + | [[image:21 9 10apo2.jpg|400px|gelfoto 21 9 10 apo2]] | ||
| + | |||
| + | |||
| + | key: JD=Jump-or-Die; CS=Cut'N'Survive; first number=Ligation number (see schedule); second number=colonie number (marked on the plate) | ||
| + | |||
| + | from left to right:7ak(short primer), 7al(long primer), JD.1a.1, JD.1a.2,JD.1b5<sub>1</sub>.1, JD.1b5<sub>2</sub>.2, JD.1b5<sub>2</sub>.1, JD.1b5<sub>2</sub>.2, JD.2.1, JD.2.2, JD.3<sub>1</sub>.1, JD.3<sub>1</sub>.2, CS.1a.1, CS.1a.2 | ||
| + | |||
| + | |||
| + | <font color="#CC33CC">'''verified products:7a (~800bp); JD.2.1; JD.2.2(each ~1000bp);JD.3<sub>1</sub>.1(~2000bp); CS.1a.2(~1500bp)'''</font> | ||
| + | |||
| + | no bands shown: CS.2b.1, CS.2b.2, PCR1.1, PCR1.CMV,3.8, 5<sub>1</sub>.7, 5<sub>2</sub>.6, 9.7, 10.5, 10.6 | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
== 9-22-2010 == | == 9-22-2010 == | ||
| - | + | <font color="#009933">Inoculate Colonies</font> | |
| + | |||
| + | inoculated in 4ml LB-medium with appropriate antibiotic | ||
| + | |||
| + | <font color="#009933">Plated residual transformated E. colis (9-20-10) (where we had few colonies on plates) </font> | ||
| + | |||
| + | <font color="#009933">Colony PCRs</font> | ||
| + | |||
| + | for 22*5µl charges: | ||
| + | |||
| + | {| | ||
| + | |PCR Mastermix | ||
| + | |55µl | ||
| + | |- | ||
| + | |Primer F | ||
| + | |8,25µl | ||
| + | |- | ||
| + | |Primer R | ||
| + | |8,25µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |38,5µl | ||
| + | |- | ||
| + | |sum | ||
| + | |110µl | ||
| + | |} | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/21 PCR with Taq Mastermix|21 PCR with Taq Mastermix]] | ||
| + | |||
| + | Annealing temperature: 53°C | ||
| + | |||
| + | |||
| + | <font color="#009933">PCR Purification of 7a</font> | ||
| + | |||
| + | charges 7ak and 7al had not been labeled, so they were renamed. | ||
| + | |||
| + | results: | ||
| + | |||
| + | 7a<sub>1</sub>: 135ng/µl; A260/A280=1.7 | ||
| + | |||
| + | 7a<sub>2</sub>: 133ng/µl; A260/A280=1.7 | ||
| + | |||
| + | |||
| + | <font color="#009933">PCR8 with Phusion Hot Start II</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/20 PCR with Phusion Hot Start|20 PCR with Phusion Hot Start]] | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |template 7a 1:100 | ||
| + | |3µl ~4ng (850bp) | ||
| + | |- | ||
| + | |template 7b 1:10 | ||
| + | |2µl ~2ng (402bp) | ||
| + | |- | ||
| + | |Buffer 5X | ||
| + | |10µl | ||
| + | |- | ||
| + | |Phusion | ||
| + | |0.5µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |27µl | ||
| + | |- | ||
| + | |Primer 13 | ||
| + | |2.5µl | ||
| + | |- | ||
| + | |Primer 16 | ||
| + | |2.55µl | ||
| + | |- | ||
| + | |DMSO) | ||
| + | |1.5µl | ||
| + | |- | ||
| + | |dNTP mix | ||
| + | |1µl | ||
| + | |} | ||
| + | |||
| + | Program: annealing temperature: 63°C, elongation time: 40sec | ||
| + | |||
| + | <font color="#009933">Agarose gel electrophoresis of Colony PCRs and PCR8</font> | ||
| + | |||
| + | new ladder: fermentas FastRuler (TM) Middle Range DNA Ladder http://www.fermentas.com/en/products/all/dna-electrophoresis/fastruler-dna-ladders/sm111-fastruler-middle | ||
| + | |||
| + | [[image: 9 22 10 beschriftet.jpg|400px|gelfoto]] | ||
| + | |||
| + | some results: | ||
| + | |||
| + | PCR 8 (lanes 23, 24): no band | ||
| + | |||
| + | Colony PCRs CS.2b.10, PCR1cam.3, PCR1cam.4, CMVcam.4, CMVcam.4, JD.1b5<sub>1</sub>.10 (lanes 7,8,9,10,11,14): show right bands | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <font color="#009933">Plasmid extraction of "over-day" cultures ligation-7(=BB4),-8(=BB4),-9(=BB5),-12(=BB9) and pSB1C3</font> | ||
| + | |||
| + | ->Protocol: [[Team:LMU-Munich/Notebook/Protocols/4 Plasmid extraction from cells|4 Plasmid extraction from cells]] | ||
| + | |||
| + | concentrations (ng/µl - A<sub>260/280</sub>): | ||
| + | ::BB4(7):57,5 - 1,643 | ||
| + | ::BB4(8):15 - 1,5 | ||
| + | ::BB5(9):173 - 1,816 | ||
| + | ::BB7(12):55,0 - 1,571 | ||
| + | ::pSB1C3: 12,5 - 1,250 | ||
| + | |||
| + | <font color="#009933">new PCR 7b in order to get better results</font> | ||
| + | {| | ||
| + | |- | ||
| + | |Master Mix | ||
| + | |25µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |16,5µl | ||
| + | |- | ||
| + | |Primer 15 | ||
| + | |3,75µl | ||
| + | |- | ||
| + | |Primer 16 | ||
| + | |3,75µl | ||
| + | |- | ||
| + | |template 190-6 (1:10) | ||
| + | |1µl | ||
| + | |} | ||
| + | |||
| + | <font color="#009933">overnight-cultures</font> | ||
| + | |||
| + | inoculated of: CS2bK10=BB11; PCP1K3+4=BB1; CMV-C-K3+4; JD1b5<sub>1</sub>K10=BB2; pSB1C3 | ||
| + | |||
== 9-23-2010 == | == 9-23-2010 == | ||
| - | + | <font color="#009933">Restriction digestion of BB5 for the 3A Method</font> | |
| + | |||
| + | {| | ||
| + | |Name | ||
| + | |H<sub>2</sub>O | ||
| + | |Buffer D | ||
| + | |BSA (1:10) | ||
| + | |DNA Volume | ||
| + | |DNA Mass | ||
| + | |Enzyms (2*0.5µl) | ||
| + | |- | ||
| + | |BB5 | ||
| + | |11 µl | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |4µl | ||
| + | |~500ng | ||
| + | |X+P | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | sum: 20µl | ||
| + | |||
| + | -> Protocol:[[Team:LMU-Munich/Notebook/Protocols/5 Restriction digest|5 Restriction digestion]] | ||
| + | |||
| + | <font color="#009933">Plasmid extraction of overnight cultures BB2, BB11, CMV in cam and pSB1C3</font> | ||
| + | |||
| + | ->Protocol: [[Team:LMU-Munich/Notebook/Protocols/4 Plasmid extraction from cells|4 Plasmid extraction from cells]] | ||
| + | |||
| + | concentrations (ng/µl - A<sub>260/280</sub>): | ||
| + | ::BB2:232 - 1,603 | ||
| + | ::BB11:170 - 1,889 | ||
| + | ::CMV in cam:272 - 1,703 | ||
| + | ::pSB1C3: 290 - 1,758 | ||
| + | |||
| + | The pSB1C3 colony was pink (produced a red pigment or something like that, so we make a new transformation in order to extract only pSB1C3 and not some other vectors. | ||
| + | |||
| + | <font color="#009933">Agarose gelelectrophoresis of PCR 7b and pathway restriction digestions</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | [[Image:23 9 10 1.jpg|thumb|right|Gelphoto]] | ||
| + | |||
| + | From left to right: PCR7b, pathway (3x),ladder (5000,2000,850,400,100bp) | ||
| + | |||
| + | result: <font color="#CC33CC">'''PCR 7b shows right band with ~400bp '''</font> | ||
| + | |||
| + | <font color="#009933">PCR clean up of PCR 7b</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/12 Gel extraction or PCR Clean up| 12 Gel extraction or PCR Clean up (Promega kit)]] | ||
| + | |||
| + | result: 90ng/µl; A260/A280= 1,895 | ||
| + | |||
| + | |||
| + | |||
| + | <font color="#009933">Restriction digestion of BB2 and BB11</font> | ||
| + | |||
| + | |||
| + | {| | ||
| + | |Name | ||
| + | |H<sub>2</sub>O | ||
| + | |Buffer D | ||
| + | |BSA (1:10) | ||
| + | |DNA Volume | ||
| + | |DNA Mass | ||
| + | |Enzyms (2*0.5µl) | ||
| + | |- | ||
| + | |BB2 | ||
| + | |10 µl | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |5µl | ||
| + | |~1.2mg | ||
| + | |X+P | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | sum: 20µl | ||
| + | |||
| + | |||
| + | {| | ||
| + | |Name | ||
| + | |H<sub>2</sub>O | ||
| + | |Buffer D | ||
| + | |BSA (1:10) | ||
| + | |DNA Volume | ||
| + | |DNA Mass | ||
| + | |Enzyms (2*0.5µl) | ||
| + | |- | ||
| + | |BB11 | ||
| + | |7 µl | ||
| + | |2µl | ||
| + | |2µl | ||
| + | |8µl | ||
| + | |~1.3mg | ||
| + | |X+P | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | sum: 20µl | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/5 Restriction digest|5 Restriction digestion]] | ||
| + | |||
| + | |||
| + | <font color="#009933">Ligation of BB1 and BB6</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/22 Ligation|22 Ligation]] | ||
| + | |||
| + | {| | ||
| + | |Name | ||
| + | |H<sub>2</sub>O | ||
| + | |T4 Ligase-Buffer | ||
| + | |PCR1 | ||
| + | |Primer 18+19 | ||
| + | |ccdBAmp | ||
| + | |T4 Ligase | ||
| + | |- | ||
| + | |BB1 | ||
| + | |9.5 µl | ||
| + | |2µl | ||
| + | |5µl | ||
| + | |2µl | ||
| + | |1µl | ||
| + | |0.5µl | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | {| | ||
| + | |Name | ||
| + | |H<sub>2</sub>O | ||
| + | |T4 Ligase-Buffer | ||
| + | |CMV E+S | ||
| + | |BB5 | ||
| + | |ccdBCam | ||
| + | |T4 Ligase | ||
| + | |- | ||
| + | |BB6 | ||
| + | |6.5 µl | ||
| + | |2µl | ||
| + | |8µl | ||
| + | |2µl | ||
| + | |1µl | ||
| + | |0.5µl | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | <font color="#009933">PCR 8 with 7b(new) plus 7a-2 with Phusion Hot Start II; Primer 16 plus 13k and 13l</font> | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |Buffer 5x | ||
| + | |10µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |27µl | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |1µl | ||
| + | |- | ||
| + | |Primer 16 | ||
| + | |2,5µl | ||
| + | |- | ||
| + | |Primer 13k short | ||
| + | |2,5µl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,5µl | ||
| + | |- | ||
| + | |template 7a(1:100)(850bp) | ||
| + | |3µl -> 4ng | ||
| + | |- | ||
| + | |template 7b(new)(1:100)(402bp) | ||
| + | |2µl -> 2ng | ||
| + | |- | ||
| + | |Phusion Hot Start II | ||
| + | |0,5µl | ||
| + | |} | ||
| + | :->same with Primer 13l long<- | ||
| + | |||
| + | <font color="#009933">Overnight culture of PCR1 in cam- Vector</font> | ||
| + | |||
| + | new inoculation of PCR1 in cam, Colony 3 in 5ml LB+cam | ||
| + | |||
| + | [[Image:23 9 10 2apo.jpg|thumb|right|From left to right: PCR8k,PCR8l,ladder (5000,2000,850,400,100bp) | ||
| + | ]] | ||
| + | |||
| + | <font color="#009933">Transformation of pSB1C3, BB1, BB6</font> | ||
| + | |||
| + | New transformation of pSB1C3 from Spring distribution because the old colony was pink. | ||
| + | |||
| + | Transformation of BB1 and BB6. | ||
| + | |||
| + | |||
| + | BB1 plated on amp-Agar, BB6 and pSB1C3 plated on cam-Agar | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/19 Transformation SSS|19 Transformation (Sina Science Services)]] | ||
| + | |||
| + | <font color="#009933">Agarose gelelectrophoresis of PCR8 (long and short primer 13)</font> | ||
| + | |||
| + | PCR8k=short primer 13, PCR8l=long primer 13 | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/11 Agarose gel electrophoresis| 11 Agarose gel electrophoresis]] | ||
| + | |||
| + | result:<font color="#CC33CC"> '''PCR 8k and 8l shows right band with ~1200bp'''</font> | ||
| + | |||
| + | <font color="#009933">PCR purification of PCR 8</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/12 Gel extraction or PCR Clean up| 12 Gel extraction or PCR Clean up (Promega kit)]] | ||
| + | |||
| + | result: ~70ng/µl; A260/A280=1.75 | ||
| + | |||
== 9-24-2010 == | == 9-24-2010 == | ||
| - | + | <font color="#009933">New "over-day" culture of pSB1C3</font> | |
| + | |||
| + | Inoculate one culture from the ligation yesterday in 4ml LB+cam | ||
| + | |||
| + | <font color="#009933">Ligation of BB7<sub>2</sub></font> | ||
| + | |||
| + | Mixture: | ||
| + | |||
| + | Inserts: each 5µl; ccdB vector: 1µl; T4 Ligase: 0.5µl; T4 Ligase Buffer: 2µl; H<sub>2</sub>O: 7µl | ||
| + | |||
| + | Incubation: 5h 18°C; Inhibition: 20 min 65°C | ||
| + | |||
| + | ->Protocols: [[Team:LMU-Munich/Notebook/Protocols/22 Ligation|22 Ligation]], [[Team:LMU-Munich/Notebook/Protocols/13 3A Method for Biobrick assembly| 13 3A Method for Biobrick assembly]] | ||
| + | |||
| + | <font color="#009933">Colony PCRs of BB1 and BB6</font> | ||
| + | |||
| + | 2 Colonys were picked from each plate -> 4 charges | ||
| + | |||
| + | Mixture for 4 charges with 5µl: | ||
| + | |||
| + | {| | ||
| + | |PCR Mastermix 2x | ||
| + | |10µl | ||
| + | |- | ||
| + | |Primer F | ||
| + | |1.5µl | ||
| + | |- | ||
| + | |Primer R | ||
| + | |1.5µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |7µl | ||
| + | |- | ||
| + | |sum | ||
| + | |20µl | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | + Colony | ||
| + | |||
| + | [[Image: apo-24.9.10.jpg|200px|thumb|right|from left to right: 1<sub>1</sub>, 1<sub>2</sub>, 6<sub>1</sub> and 6<sub>2</sub>]] | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/21 PCR with Taq Mastermix|21 PCR with Taq Mastermix]] | ||
| + | |||
| + | Annealing temperature: 53°C, standard protocol | ||
| + | |||
| + | <font color="#009933">Plasmid extraction of new PCR1 in cam vector</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/4 Plasmid extraction from cells|4 Plasmid extraction from cells]] | ||
| + | |||
| + | results: | ||
| + | |||
| + | {| | ||
| + | | concentration | ||
| + | | 82.5ng/µl | ||
| + | |- | ||
| + | | A260/A280 | ||
| + | | 1.737 | ||
| + | |} | ||
| + | |||
| + | |||
| + | <font color="#009933">Agarose Gelelektrophoresis of 1<sub>1</sub>, 1<sub>2</sub>, 6<sub>1</sub> and 6<sub>2</sub></font> | ||
| + | |||
| + | |||
| + | <font color="#009933">Plasmid extraction of pSB1C3 in cam vector</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/4 Plasmid extraction from cells|4 Plasmid extraction from cells]] | ||
| + | |||
| + | results: | ||
| + | |||
| + | {| | ||
| + | | concentration | ||
| + | | 22.5ng/µl | ||
| + | |- | ||
| + | | A260/A280 | ||
| + | | 1.5 | ||
| + | |} | ||
| + | |||
== 9-25-2010 == | == 9-25-2010 == | ||
weekend | weekend | ||
| Line 672: | Line 4,072: | ||
== 9-27-2010 == | == 9-27-2010 == | ||
| - | + | ||
| + | <font color="#009933">Colony PCR of BB7<sub>2</sub>, BB6 and BB1 plus a negativ charge as reference</font> | ||
| + | |||
| + | {| | ||
| + | | PCR Mastermix | ||
| + | | 17.5µl | ||
| + | |- | ||
| + | | Primer forward | ||
| + | | 2.625µl | ||
| + | |- | ||
| + | | Primer revers | ||
| + | | 2.7µl | ||
| + | |- | ||
| + | | H<sub>2</sub>O | ||
| + | | 12.1µl | ||
| + | |- | ||
| + | | sum | ||
| + | | 35µl = 5µl per charge | ||
| + | |} | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/24 Colony PCR|24 Colony PCR]] | ||
| + | |||
| + | |||
| + | <font color="#009933">Ligation of BB10</font> | ||
| + | |||
| + | {| | ||
| + | | CMV© E+S | ||
| + | | 5µl | ||
| + | |- | ||
| + | | PCR 8l X+P | ||
| + | | 5µl | ||
| + | |- | ||
| + | | ccdBamp | ||
| + | | 1µl | ||
| + | |- | ||
| + | | T4 Ligase-Buffer | ||
| + | | 2µl | ||
| + | |- | ||
| + | | T4 Ligase | ||
| + | | 0.5µl | ||
| + | |- | ||
| + | | H<sub>2</sub>O | ||
| + | | 6.5µl | ||
| + | |- | ||
| + | | sum | ||
| + | | 20µl | ||
| + | |} | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/22 Ligation|22 Ligation]] | ||
| + | |||
| + | |||
| + | <font color="#009933">Ligation of BB001(PCR1 in ©), BB017(PCR5 in ©), BB018(PCR6 in ©) and BB021(PCR9 in ©)</font> | ||
| + | |||
| + | {| | ||
| + | | ccdB | ||
| + | | 1µl | ||
| + | |- | ||
| + | | BB001/017/018/021 | ||
| + | | 5µl | ||
| + | |- | ||
| + | | T4 Ligase-Buffer | ||
| + | | 2µl | ||
| + | |- | ||
| + | | T4 Ligase | ||
| + | | 0.5µl | ||
| + | |- | ||
| + | | H<sub>2</sub>O | ||
| + | | 11.5µl | ||
| + | |- | ||
| + | | sum | ||
| + | | 20µl | ||
| + | |} | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/22 Ligation|22 Ligation]] | ||
| + | |||
| + | <font color="#009933">Restriction digestion of PCR3<sub>2</sub>, 8l, 10 and Primer 18+19 with E+P</font> | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! | ||
| + | ! template | ||
| + | ! H<sub>2</sub>O | ||
| + | ! BSA [1:10] | ||
| + | ! Buffer H | ||
| + | ! EcoRI | ||
| + | ! PstI | ||
| + | |- | ||
| + | | PCR3<sub>2</sub> | ||
| + | | 10µl | ||
| + | | 5µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 0.5µl | ||
| + | | 0.5µl | ||
| + | |- | ||
| + | | PCR8l | ||
| + | | 10µl | ||
| + | | 5µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 0.5µl | ||
| + | | 0.5µl | ||
| + | |- | ||
| + | | Primer 18+19 | ||
| + | | 2x7.5µl | ||
| + | | -- | ||
| + | |2µl | ||
| + | | 2µl | ||
| + | | 0.5µl | ||
| + | | 0.5µl | ||
| + | |- | ||
| + | | PCR10 | ||
| + | | 10µl | ||
| + | | 5µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 0.5µl | ||
| + | | 0.5µl | ||
| + | |} | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/5 Restriction digest|5 Restriction digestion]] | ||
| + | |||
== 9-28-2010 == | == 9-28-2010 == | ||
| - | + | <font color="#009933">Plating BB017, BB021, BB018, BB001, BB7<sub>2</sub>, BB10</font> | |
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | | Amp | ||
| + | | BB7<sub>2</sub> | ||
| + | | BB10 | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | | Cam | ||
| + | | BB001 | ||
| + | | BB017 | ||
| + | | BB018 | ||
| + | | BB021 | ||
| + | |} | ||
| + | |||
| + | <font color="#009933">Colony PCR BB1, BB6, BB7<sub>2</sub> with DreamTaq (each 2 colonies)</font> | ||
| + | |||
| + | -> Protocol: [[Team:LMU-Munich/Notebook/Protocols/16 PCR with DreamTaq| 16 PCR with DreamTaq]] | ||
| + | |||
| + | PCR mixture for PCR7a | ||
| + | ::{| | ||
| + | |- | ||
| + | |template | ||
| + | |pick up colony from plate | ||
| + | |- | ||
| + | |primers*2 | ||
| + | |0.25µl*2 | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |0.5µl | ||
| + | |- | ||
| + | |DreamTaq | ||
| + | |0.05µl | ||
| + | |- | ||
| + | |10xbuffer | ||
| + | |0.5µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |3.45µl | ||
| + | |- | ||
| + | |sum | ||
| + | |5µl | ||
| + | |} | ||
| + | |||
| + | Primers for colony PCR: BBseqF, BBseqR | ||
| + | |||
| + | Annealing temperature: 53°C | ||
| + | |||
| + | <font color="#009933">Ligation of BB2, BB019, BB020, BB016, BB022</font> | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! | ||
| + | ! BB2 | ||
| + | ! BB019 | ||
| + | ! BB020 | ||
| + | ! BB016 | ||
| + | ! BB022 | ||
| + | ! Volume | ||
| + | |- | ||
| + | | PCR | ||
| + | | 5<sub>2</sub> | ||
| + | | 8 | ||
| + | | | ||
| + | | 3<sub>2</sub> | ||
| + | | 10 | ||
| + | | each 5µl | ||
| + | |- | ||
| + | | PCR | ||
| + | | 6 | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | each 5µl | ||
| + | |- | ||
| + | | Primer | ||
| + | | | ||
| + | | | ||
| + | | 18+19 | ||
| + | | | ||
| + | | | ||
| + | | each 5µl | ||
| + | |- | ||
| + | | ccdBamp | ||
| + | | 1µl | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | | ccdBcam | ||
| + | | | ||
| + | | 1µl | ||
| + | | 1µl | ||
| + | | 1µl | ||
| + | | 1µl | ||
| + | | | ||
| + | |- | ||
| + | | T4 Ligase-Buffer | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | | ||
| + | |- | ||
| + | | T4 Ligase | ||
| + | | 0.5µl | ||
| + | | 0.5µl | ||
| + | | 0.5µl | ||
| + | | 0.5µl | ||
| + | | 0.5µl | ||
| + | | | ||
| + | |- | ||
| + | | H<sub>2</sub>O | ||
| + | | 6.5µl | ||
| + | | 11.4µl | ||
| + | | 11.4µl | ||
| + | | 11.4µl | ||
| + | | 11.4µl | ||
| + | | | ||
| + | |- | ||
| + | | sum | ||
| + | | 20µl | ||
| + | | 20µl | ||
| + | | 20µl | ||
| + | | 20µl | ||
| + | | 20µl | ||
| + | | | ||
| + | |} | ||
| + | |||
== 9-29-2010 == | == 9-29-2010 == | ||
| - | + | plates BB7<sub>2</sub>, BB017, BB10, BB021, BB001, BB018: nothing grown -> thrown into trash | |
| + | |||
| + | <font color="#009933">colony-PCR of BB2/8, BB019, BB016, BB020, BB022 with Taq</font> | ||
| + | |||
| + | ->protocol: [[Team:LMU-Munich/Notebook/Protocols/25 PCR with Taq|PCR with Taq]] | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |6,7µl | ||
| + | |- | ||
| + | |Buffer 10x | ||
| + | |1µl | ||
| + | |- | ||
| + | |MgCl<sub>2</sub> | ||
| + | |1µl | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |0,2µl | ||
| + | |- | ||
| + | |Primer forw. seq f | ||
| + | |0,5µl | ||
| + | |- | ||
| + | |Primer rev. seq r | ||
| + | |0,5µl | ||
| + | |- | ||
| + | |Taq | ||
| + | |0,1µl | ||
| + | |} | ||
| + | |||
| + | template: pick colony | ||
| + | |||
| + | sum: 10µl; we made 100µl ->10 charges (2 colonies of each plate) | ||
| + | |||
| + | tube numbers: | ||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | |1 | ||
| + | |2 | ||
| + | |3 | ||
| + | |4 | ||
| + | |5 | ||
| + | |6 | ||
| + | |7 | ||
| + | |8 | ||
| + | |9 | ||
| + | |10 | ||
| + | |- | ||
| + | |BB2/BB8 - 1 | ||
| + | |BB2/BB8 - 2 | ||
| + | |BB016 - 1 | ||
| + | |BB016 - 2 | ||
| + | |BB019 - 1 | ||
| + | |BB019 - 2 | ||
| + | |BB020 - 1 | ||
| + | |BB020 - 2 | ||
| + | |BB022 - 1 | ||
| + | |BB022 - 2 | ||
| + | |} | ||
| + | |||
| + | PCR-program: taq53 | ||
| + | |||
| + | agarose-gelelektrophoresis -> no bands visible | ||
| + | |||
| + | <font color="#009933">new plates made of BB1, BB4, BB6, BB7</font> | ||
| + | |||
| + | <font color="#009933">Ligations BB7<sub>2</sub>, BB017 (mit PCR5<sub>1</sub>), BB10, BB021, BB001, BB018</font> | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | | | ||
| + | |PCR (5 µl each) | ||
| + | |PCR (5µl each) | ||
| + | |ccdB(amp) | ||
| + | |ccdB© | ||
| + | |Buffer | ||
| + | |T4 Ligase | ||
| + | |H<sub>2</sub>O | ||
| + | |- | ||
| + | |BB7<sub>2</sub>O | ||
| + | |1 | ||
| + | |3<sub>2</sub> | ||
| + | |1µl | ||
| + | | | ||
| + | |2µl | ||
| + | |0,5µl | ||
| + | |6,5µl | ||
| + | |- | ||
| + | |BB017 | ||
| + | |5<sub>1</sub> | ||
| + | | | ||
| + | | | ||
| + | |1µl | ||
| + | |2µl | ||
| + | |0,5µl | ||
| + | |11,5µl | ||
| + | |- | ||
| + | |BB10 | ||
| + | |CMV© | ||
| + | |PCR8 | ||
| + | |1µl | ||
| + | | | ||
| + | |2µl | ||
| + | |0,5µl | ||
| + | |6,5µl | ||
| + | |- | ||
| + | |BB021 | ||
| + | |9 | ||
| + | | | ||
| + | | | ||
| + | |1µl | ||
| + | |2µl | ||
| + | |0,5µl | ||
| + | |11,5µl | ||
| + | |- | ||
| + | |BB001 | ||
| + | |1 | ||
| + | | | ||
| + | | | ||
| + | |1µl | ||
| + | |2µl | ||
| + | |0,5µl | ||
| + | |11,5µl | ||
| + | |- | ||
| + | |BB018 | ||
| + | |6 | ||
| + | | | ||
| + | | | ||
| + | |1µl | ||
| + | |2µl | ||
| + | |0,5µl | ||
| + | |11,5µl | ||
| + | |} | ||
| + | |||
| + | <font color="#009933">restriction degestions</font> | ||
| + | |||
| + | of PCR6, PCR5<sub>2</sub>, PCR3<sub>2</sub>, PCR1, ccdB(amp), ccdB(cam) | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | |template | ||
| + | |digestion with | ||
| + | |volume | ||
| + | |- | ||
| + | |PCR 6 | ||
| + | |Xba1 & Pst1 | ||
| + | |I. 10µl II. 3µl | ||
| + | |- | ||
| + | |PCR5<sub>2</sub> | ||
| + | |EcoR1 & Pst1 | ||
| + | |20µl | ||
| + | |- | ||
| + | |PCR3<sub>2</sub> | ||
| + | |Xba1 & Pst1 | ||
| + | |10µl | ||
| + | |- | ||
| + | |PCR1 | ||
| + | |EcoR1 & Spe1 | ||
| + | |2µl | ||
| + | |- | ||
| + | |ccdB(amp) | ||
| + | |EcoR1 & Pst1 | ||
| + | |20µl | ||
| + | |- | ||
| + | |cddB(cam) | ||
| + | |EcoR1 & Pst1 | ||
| + | |20µl | ||
| + | |} | ||
| + | ->protocol: [[Team:LMU-Munich/Notebook/Protocols/5 Restriction digest|5 Restriction digestion]] | ||
| + | |||
| + | |||
| + | <font color="#009933">colony-PCR of BB4/c3, BB6/c7, BB7<sub>2</sub>/c5, BB1/c5, (BB11 as control) with Taq</font> | ||
| + | |||
| + | ->protocol: [[Team:LMU-Munich/Notebook/Protocols/25 PCR with Taq|PCR with Taq]] | ||
| + | |||
| + | Mastermix: | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |32µl | ||
| + | |- | ||
| + | |Buffer 10x | ||
| + | |5µl | ||
| + | |- | ||
| + | |MgCl<sub>2</sub> | ||
| + | |5µl | ||
| + | |- | ||
| + | |dNTPs | ||
| + | |1µl | ||
| + | |- | ||
| + | |Primer forw. seq f | ||
| + | |2,5µl | ||
| + | |- | ||
| + | |Primer rev. seq r | ||
| + | |2,5µl | ||
| + | |- | ||
| + | |Taq | ||
| + | |0,5µl | ||
| + | |- | ||
| + | |DMSO | ||
| + | |1,5µl | ||
| + | |- | ||
| + | |sum | ||
| + | |50µl | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | ->10µl for each charge | ||
| + | |||
| + | template: pick colony; BB11: 2ng | ||
| + | |||
| + | tube numbers: | ||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | |1 | ||
| + | |2 | ||
| + | |3 | ||
| + | |4 | ||
| + | |5 | ||
| + | |- | ||
| + | |BB4-c3 | ||
| + | |BB6-c7 | ||
| + | |BB7<sub>2</sub>-c5 | ||
| + | |BB1-c5 | ||
| + | |BB11 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | PCR program:T<sub>A</sub> = 53°C; elongation time=1min | ||
| + | |||
== 9-30-2010 == | == 9-30-2010 == | ||
| - | + | <font color="#009933">Colony PCR of BB2/BB8, BB6 K7&K8 and test PCR of BB11 with new dilutions and primers</font> | |
| + | |||
| + | {| | ||
| + | | Colony/BB11 | ||
| + | | 2.5µl | ||
| + | |- | ||
| + | | H<sub>2</sub>O | ||
| + | | 32µl | ||
| + | |- | ||
| + | | dNTP | ||
| + | | 1µl | ||
| + | |- | ||
| + | | Primer F | ||
| + | | 2.5µl | ||
| + | |- | ||
| + | | Primer R | ||
| + | | 2.5µl | ||
| + | |- | ||
| + | | MgCl<sub>2</sub> | ||
| + | | 5µl | ||
| + | |- | ||
| + | | DMSO | ||
| + | | 1.5µl | ||
| + | |- | ||
| + | | Buffer 10x (with KCl) | ||
| + | | 5µl | ||
| + | |- | ||
| + | | Taq Polymerase | ||
| + | | 0.5µl | ||
| + | |- | ||
| + | | sum | ||
| + | | 56µl | ||
| + | |} | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/24 Colony PCR|24 Colony PCR]] | ||
| + | |||
| + | <font color="#009933">PCR purification of the digestions of 6I, 6II, 5<sub>2</sub>, 3<sub>2</sub>, 1</font> | ||
| + | |||
| + | {| | ||
| + | | | ||
| + | | concentration [ng/µl] | ||
| + | | A260/A280 | ||
| + | |- | ||
| + | | 6II | ||
| + | | 32.5 | ||
| + | | 1.444 | ||
| + | |- | ||
| + | | ccdBamp | ||
| + | | 0.0 | ||
| + | | --- | ||
| + | |- | ||
| + | | ccdBcam | ||
| + | | 5.0 | ||
| + | | 1.000 | ||
| + | |- | ||
| + | | 6I | ||
| + | | 0.0 | ||
| + | | --- | ||
| + | |- | ||
| + | | 5<sub>2</sub> | ||
| + | | 20.0 | ||
| + | | 1.143 | ||
| + | |- | ||
| + | | 3<sub>2</sub> | ||
| + | | 27.5 | ||
| + | | 1.57 | ||
| + | |- | ||
| + | | 1 | ||
| + | | 25 | ||
| + | | 1.667 | ||
| + | |} | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/12 Gel extraction or PCR Clean up| 12 Gel extraction or PCR Clean up (Promega kit)]] | ||
| + | |||
| + | <font color="#009933">Transformation of BB018, BB7<sub>2</sub>, BB001, BB017, BB10 and BB021</font> | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/19 Transformation SSS|19 Transformation SSS]] | ||
| + | |||
| + | <font color="#009933">Plasmidextraction of the following overnight cultures: BB020(K1), BB018(K5), BB6(K5), BB022(K1), BB016(K1), BB019(K1), BB2(K1) and BB018(K3)</font> | ||
| + | |||
| + | {| | ||
| + | | | ||
| + | | concentration [ng/µl] | ||
| + | | A260/A280 | ||
| + | |- | ||
| + | | BB020(K1) | ||
| + | | 163 | ||
| + | | 1.711 | ||
| + | |- | ||
| + | | BB018(K5) | ||
| + | | 92.5 | ||
| + | | 1.85 | ||
| + | |- | ||
| + | | BB6(K5) | ||
| + | | 120 | ||
| + | | 1.778 | ||
| + | |- | ||
| + | | BB022(K1) | ||
| + | | 105 | ||
| + | | 1.75 | ||
| + | |- | ||
| + | | BB016(K1) | ||
| + | | 85 | ||
| + | | 1.889 | ||
| + | |- | ||
| + | | BB019(K1) | ||
| + | | 105 | ||
| + | | 1.909 | ||
| + | |- | ||
| + | | BB2(K1) | ||
| + | | 190 | ||
| + | | 1.81 | ||
| + | |- | ||
| + | | BB018 | ||
| + | | 110 | ||
| + | | 1.833 | ||
| + | |} | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/4 Plasmid extraction from cells|4 Plasmid extraction from cells]] | ||
| + | |||
== 10-01-2010 == | == 10-01-2010 == | ||
| - | + | <font color="#009933">new test PCR of BB5 and BB11 with 20µl and 50µl volume</font> | |
| + | |||
| + | Master Mix | ||
| + | {| | ||
| + | | H<sub>2</sub>O | ||
| + | | 67µl | ||
| + | |- | ||
| + | | dNTP | ||
| + | | 2µl | ||
| + | |- | ||
| + | | Primer F | ||
| + | | 5µl | ||
| + | |- | ||
| + | | Primer R | ||
| + | | 5µl | ||
| + | |- | ||
| + | | MgCl<sub>2</sub> | ||
| + | | 10µl | ||
| + | |- | ||
| + | | Buffer 10x (with KCl) | ||
| + | | 10µl | ||
| + | |- | ||
| + | | Taq Polymerase | ||
| + | | 1µl | ||
| + | |- | ||
| + | | sum | ||
| + | | 100µl | ||
| + | |} | ||
| + | |||
| + | template: BB5/50µl: 5ng, 20µl: 1.5ng; BB11/20µl: 3ng | ||
| + | |||
| + | ->protocol: [[Team:LMU-Munich/Notebook/Protocols/25 PCR with Taq|PCR with Taq]] | ||
| + | |||
| + | [[Image:01.10.10. apo.jpg|01.10.10. Gel photo]] | ||
| + | |||
| + | from left to right: ladder, BB5/50µl, BB5/20µl, BB11/20µl. BB5/50µl and BB11/20µl show the right band. | ||
| + | |||
== 10-02-2010 == | == 10-02-2010 == | ||
weekend | weekend | ||
== 10-03-2010 == | == 10-03-2010 == | ||
weekend | weekend | ||
| + | |||
| + | == 10-04-2010 == | ||
| + | sequencing | ||
| + | |||
| + | cycle, clean and run | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! | ||
| + | ! CMV (c) | ||
| + | ! PCR 1 (c) | ||
| + | ! BB4 <sub>7</sub> | ||
| + | ! BB4 <sub>8</sub> | ||
| + | ! BB018K5 | ||
| + | ! BB018K3 | ||
| + | ! BB6K5 | ||
| + | ! BB2K1 | ||
| + | |- | ||
| + | | template | ||
| + | | 1µl | ||
| + | | 2µl | ||
| + | | 3µl | ||
| + | | 3,8µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 1µl | ||
| + | |- | ||
| + | | primer (1:100) | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | |- | ||
| + | | 10mM TrispH 8,5 | ||
| + | | 2,8 µl | ||
| + | | 1,8 µl | ||
| + | | 0,8 µl | ||
| + | | 0 µl | ||
| + | | 1,8 µl | ||
| + | | 1,8 µl | ||
| + | | 1,8 µl | ||
| + | | 1,8 µl | ||
| + | |- | ||
| + | |name of tube | ||
| + | | 028 FIR | ||
| + | | 001 F/R | ||
| + | | 4-7 F/R | ||
| + | | 4-8 F/R | ||
| + | | 018-5 F/R | ||
| + | | 018-3 F/R | ||
| + | | 6-5 F/R | ||
| + | | 2-1 F/R | ||
| + | |} | ||
| + | |||
| + | |||
| + | PCR | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! | ||
| + | ! 019 | ||
| + | ! 022 | ||
| + | ! 016 | ||
| + | ! 020 | ||
| + | |- | ||
| + | | template (1:100 = 2ng) | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 2,5µl | ||
| + | | 1,5µl | ||
| + | |- | ||
| + | | Primer F | ||
| + | | 1µl | ||
| + | | 1µl | ||
| + | | 1µl | ||
| + | | 1µl | ||
| + | |- | ||
| + | | Primer R | ||
| + | | 1µl | ||
| + | | 1µl | ||
| + | | 1µl | ||
| + | | 1µl | ||
| + | |- | ||
| + | | dNTPs | ||
| + | | 0,4µl | ||
| + | | 0,4µl | ||
| + | | 0,4µl | ||
| + | | 0,4µl | ||
| + | |- | ||
| + | | Taq | ||
| + | | 0,2µl | ||
| + | | 0,2µl | ||
| + | | 0,2µl | ||
| + | | 0,2µl | ||
| + | |- | ||
| + | | Buffer | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | |- | ||
| + | |MgCl<sub>2</sub> | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | | 11,4µl | ||
| + | | 11,4µl | ||
| + | | 10,9µl | ||
| + | | 11, | ||
| + | |- | ||
| + | |colour of tube | ||
| + | | orange | ||
| + | | rose | ||
| + | | green | ||
| + | | blue | ||
| + | |} | ||
| + | |||
| + | == 10-05-2010 == | ||
| + | <font color="#009933">Colony PCRs of BB6, BB4, BB021, BB022, CMV©, BB001, BB016, BB1, BB2<sub>5<sub>2</sub></sub>, BB7<sub>2</sub>, BB017, BB018, BB019, BB020</font> | ||
| + | |||
| + | <font color="#009933">agarose gel electrophoresis</font> | ||
| + | |||
| + | schedule of the agarose gel electrophoresis: | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | | gel 1 | ||
| + | | BB6K9 | ||
| + | | BB6K10 | ||
| + | | BB6K11 | ||
| + | | BBK12 | ||
| + | | BB6K13 | ||
| + | | BB4K5 | ||
| + | | ladder | ||
| + | | BB4K6 | ||
| + | | BB4K7 | ||
| + | |- | ||
| + | | | ||
| + | | BB021K14 | ||
| + | | BB021K13 | ||
| + | | BB021K12 | ||
| + | | BB021K11 | ||
| + | | BB022K18 | ||
| + | | BB022K19 | ||
| + | | ladder | ||
| + | | BB022K17 | ||
| + | | BB022K16 | ||
| + | |- | ||
| + | | gel 2 | ||
| + | | BB022K12 | ||
| + | | BB022K11 | ||
| + | | CMV©K4 | ||
| + | | CMV©K5 | ||
| + | | BB001K5 | ||
| + | | BB001K6 | ||
| + | | BB001K7 | ||
| + | | BB001K8 | ||
| + | | ladder | ||
| + | |- | ||
| + | | | ||
| + | | BB016K17 | ||
| + | | BB1K16 | ||
| + | | BB1K15 | ||
| + | | BB1K8 | ||
| + | | BB1K7 | ||
| + | | BB2<sub>5<sub>2</sub></sub>K1 | ||
| + | | BB2<sub>5<sub>2</sub></sub>K2 | ||
| + | | BB2<sub>5<sub>2</sub></sub>K3 | ||
| + | | ladder | ||
| + | |- | ||
| + | | gel 3 | ||
| + | | BB017K14 | ||
| + | | BB017K13 | ||
| + | | BB017K12 | ||
| + | | BB017K11 | ||
| + | | BB018K17 | ||
| + | | BB018K16 | ||
| + | | BB018K15 | ||
| + | | BB018K14 | ||
| + | | ladder | ||
| + | |- | ||
| + | | | ||
| + | | BB019K6 | ||
| + | | BB019K5 | ||
| + | | BB019K4 | ||
| + | | ladder | ||
| + | | BB019K3 | ||
| + | | BB020K7 | ||
| + | | BB020K8 | ||
| + | | BB020K6 | ||
| + | | BB020K5 | ||
| + | |} | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | | gel 1 | ||
| + | | BB4K8 | ||
| + | | BB4K9 | ||
| + | | BB4K10 | ||
| + | | BB021K10 | ||
| + | | BB021K15 | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | | | ||
| + | | BB022K15 | ||
| + | | BB022K14 | ||
| + | | BB022K13 | ||
| + | | --- | ||
| + | | --- | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | | gel 2 | ||
| + | | BB001K9 | ||
| + | | BB001K10 | ||
| + | | BB016K11 | ||
| + | | BB016K12 | ||
| + | | BB016K13 | ||
| + | | BB016K14 | ||
| + | | BB016K15 | ||
| + | | BB016K16 | ||
| + | | --- | ||
| + | |- | ||
| + | | | ||
| + | | --- | ||
| + | | BB2<sub>5<sub>2</sub></sub>K4 | ||
| + | | BB2<sub>5<sub>2</sub></sub>K5 | ||
| + | | BB7<sub>2</sub>K12 | ||
| + | | BB7<sub>2</sub>K11 | ||
| + | | BB7<sub>2</sub>K10 | ||
| + | | BB7<sub>2</sub>K9 | ||
| + | | BB017K16 | ||
| + | | BB017K15 | ||
| + | |- | ||
| + | |gel 3 | ||
| + | | BB018K13 | ||
| + | | BB018K12 | ||
| + | | BB018K11 | ||
| + | | BB018K10 | ||
| + | | BB018K9 | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | | | ||
| + | | BB020K4 | ||
| + | | BB020K3 | ||
| + | | --- | ||
| + | | --- | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | Gelbilder und Auswertung einfügen | ||
| + | |||
| + | [[Image:5.10.10apo1,2,3.jpg|400px|9 15 10 Gelphoto]] | ||
| + | |||
| + | |||
| + | |||
| + | <font color="#009933">overnight cultures of BB6K3, BB4K10, CMV©K4, BB001K5, BB001K9, BB016K11, BB1K8, BB2<sub>5<sub>2</sub></sub>K3, BB7<sub>2</sub>, BB017K14, BB018K10, BB019K6</font> | ||
| + | |||
| + | == 10-06-2010 == | ||
| + | |||
| + | <font color="#009933">Plasmidextraction of the following overnight cultures: BB6(K13), BB4(K13), CMVc (K4), BB001(K5), BB001(K9), BB016(K11), BB1(K8), BB2(K3), BB7(K10), BB017(K14), BB018(K10) and BB019(K6) </font> | ||
| + | |||
| + | {| | ||
| + | | | ||
| + | | concentration [ng/µl] | ||
| + | | A260/A280 | ||
| + | |- | ||
| + | | BB6(K13) | ||
| + | | 265 | ||
| + | | 1.8 | ||
| + | |- | ||
| + | | BB4(K10) | ||
| + | | 40 | ||
| + | | 2.0 | ||
| + | |- | ||
| + | | CMVc(K4) | ||
| + | | 97.5 | ||
| + | | 1.8 | ||
| + | |- | ||
| + | | BB001(K5) | ||
| + | | 30 | ||
| + | | 1.7 | ||
| + | |- | ||
| + | | BB001(K9) | ||
| + | | 17.5 | ||
| + | | 1.7 | ||
| + | |- | ||
| + | | BB016(K11) | ||
| + | | 45 | ||
| + | | 1.7 | ||
| + | |- | ||
| + | | BB1(K8) | ||
| + | | 57.5 | ||
| + | | 1.9 | ||
| + | |- | ||
| + | | BB2(K3) | ||
| + | | 60 | ||
| + | | 1.8 | ||
| + | |- | ||
| + | | BB7(K10) | ||
| + | | 30 | ||
| + | | 1.7 | ||
| + | |- | ||
| + | | BB017(K14) | ||
| + | | 12.5 | ||
| + | | 1.5 | ||
| + | |- | ||
| + | | BB018(K10) | ||
| + | | 35 | ||
| + | | 1.8 | ||
| + | |- | ||
| + | |BB019(K6) | ||
| + | | 37.5 | ||
| + | | 1.5 | ||
| + | |- | ||
| + | |||
| + | |} | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/4 Plasmid extraction from cells|4 Plasmid extraction from cells]] | ||
| + | |||
| + | == 10-07-2010 == | ||
| + | |||
| + | <font color="#009933"> colony PCR of BB021, BB022 with Taq (53°C) </font> | ||
| + | |||
| + | |||
| + | |||
| + | {| | ||
| + | | Primer fw | ||
| + | | 0.5 µl | ||
| + | |- | ||
| + | | Primer rev | ||
| + | | 0.5 µl | ||
| + | |- | ||
| + | | dNTPs | ||
| + | | 0.2 µl | ||
| + | |- | ||
| + | | Taq | ||
| + | | 0.1 µl | ||
| + | |- | ||
| + | | 10xbuffer | ||
| + | | 1 µl | ||
| + | |- | ||
| + | | MgCl<sub>2</sub> | ||
| + | | 1 µl | ||
| + | |- | ||
| + | | H<sub>2</sub>O | ||
| + | | 6.7 µl | ||
| + | |- | ||
| + | |||
| + | |} | ||
| + | |||
| + | == 10-08-2010 == | ||
| + | |||
| + | |||
| + | <font color="#009933"> | ||
| + | plasmide extractions from 10-06-2010 were put in a lyophilizer and redissolved in less water (10-20 µl) | ||
| + | </font> | ||
| + | {| | ||
| + | | | ||
| + | | concentration [ng/µl] | ||
| + | | A260/A280 | ||
| + | |- | ||
| + | | BB4(K10) | ||
| + | | 258 | ||
| + | | 1,981 | ||
| + | |- | ||
| + | | BB001(K5) | ||
| + | | 325 | ||
| + | | 1.94 | ||
| + | |- | ||
| + | | BB001(K9) | ||
| + | | 205 | ||
| + | | 1.907 | ||
| + | |- | ||
| + | | BB7(K10) | ||
| + | | 170 | ||
| + | | 1.744 | ||
| + | |- | ||
| + | | BB017(K14) | ||
| + | | 215 | ||
| + | | 1.955 | ||
| + | |- | ||
| + | | BB018(K10) | ||
| + | | 500 | ||
| + | | 1.88 | ||
| + | |- | ||
| + | |BB019(K6) | ||
| + | | 293 | ||
| + | | 1.95 | ||
| + | |- | ||
| + | |||
| + | |} | ||
| + | |||
| + | == 10-09-2010 == | ||
| + | <font color="#009933">Plasmid extraction BB018K5, BB018K6, BB021K17 and BB021K24</font color> | ||
| + | |||
| + | concentration of these samples to be measured on Monday | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/4 Plasmid extraction from cells|4 Plasmid extraction from cells]] | ||
| + | |||
| + | == 10-10-2010 == | ||
| + | weekend | ||
| + | |||
| + | == 10-11-2010 == | ||
| + | |||
| + | |||
| + | <font color="#009933">Plasmid extraction BB018K5, BB018K6, BB021K24 and BB021K17 from 10-09-2010 | ||
| + | </font> | ||
| + | |||
| + | |||
| + | {| | ||
| + | | | ||
| + | | concentration [ng/µl] | ||
| + | | A260/A280 | ||
| + | |- | ||
| + | | BB018K5 | ||
| + | | 55 | ||
| + | | 1.571 | ||
| + | |- | ||
| + | | BB018K6 | ||
| + | | 92,5 | ||
| + | | 1.762 | ||
| + | |- | ||
| + | | BB021K24 | ||
| + | | 27.5 | ||
| + | | 1.833 | ||
| + | |- | ||
| + | | BB021K17 | ||
| + | | 37.5 | ||
| + | | 1.508 | ||
| + | |- | ||
| + | |||
| + | |} | ||
| + | |||
| + | |||
| + | <font color="#009933">Colony PCR BB01, BB018, BB6, BB252</font> | ||
| + | |||
| + | {| | ||
| + | | stripe 1 | ||
| + | | BB001K5 | ||
| + | | K6 | ||
| + | | K7 | ||
| + | | K8 | ||
| + | | K9 | ||
| + | | K9 | ||
| + | | K10 | ||
| + | | BB252 K2 | ||
| + | |- | ||
| + | | stripe 2 | ||
| + | | BB252 K3 | ||
| + | | K4 | ||
| + | | K5 | ||
| + | | BB6 K9 | ||
| + | | K10 | ||
| + | | K11 | ||
| + | | K12 | ||
| + | | K13 | ||
| + | |- | ||
| + | | stripe 3 | ||
| + | | BB018 K9 | ||
| + | | K10 | ||
| + | | K11 | ||
| + | | K12 | ||
| + | | K13 | ||
| + | | K14 | ||
| + | | K15 | ||
| + | | K16 | ||
| + | |- | ||
| + | |single sample | ||
| + | |BB018 K17 | ||
| + | |- | ||
| + | |||
| + | |} | ||
| + | |||
| + | |||
| + | gel picture colony PCR | ||
| + | |||
| + | foto einfügen! | ||
| + | |||
| + | <font color="#009933">Sequencing 8,9 and 10 (cycle, clean and run)</font> | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! | ||
| + | ! BB001K9 for | ||
| + | ! BB001K9 rev | ||
| + | ! BB4K16 for | ||
| + | ! BB4K16 rev | ||
| + | ! BB001K5 for | ||
| + | ! BB001K5 rev | ||
| + | ! BB2<sub>5<sub>2</sub></sub>K3 | ||
| + | ! BB2<sub>5<sub>2</sub></sub>K3 | ||
| + | |- | ||
| + | | template | ||
| + | | 1µl | ||
| + | | 1µl | ||
| + | | 1µl | ||
| + | | 1µl | ||
| + | | 0,5µl | ||
| + | | 0,5µl | ||
| + | | 3,8µl | ||
| + | | 3,8µl | ||
| + | |- | ||
| + | | primer (1:100) | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | |- | ||
| + | | 10mM TrispH 8,5 | ||
| + | | 2,8 µl | ||
| + | | 2,8 µl | ||
| + | | 2,8 µl | ||
| + | | 2,8 µl | ||
| + | | 3,3 µl | ||
| + | | 3,3 µl | ||
| + | | 0 µl | ||
| + | | 0 µl | ||
| + | |- | ||
| + | |name of tube | ||
| + | | 31-F | ||
| + | | 31-R | ||
| + | | 44-F | ||
| + | | 44-R | ||
| + | | 24-F | ||
| + | | 24-R | ||
| + | | 32-F | ||
| + | | 32-R | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! | ||
| + | ! BB7<sub>2</sub>K10 for | ||
| + | ! BB7<sub>2</sub>K10 rev | ||
| + | ! BB017K14 for | ||
| + | ! BB017K14 rev | ||
| + | ! BB018K10 for | ||
| + | ! BB018K10 rev | ||
| + | ! BB019K6 for | ||
| + | ! BB019K6 rev | ||
| + | |- | ||
| + | | template | ||
| + | | 1,5µl | ||
| + | | 1,5µl | ||
| + | | 1µl | ||
| + | | 1µl | ||
| + | | 0,5µl | ||
| + | | 0,5µl | ||
| + | | 0,8µl | ||
| + | | 0,8µl | ||
| + | |- | ||
| + | | primer (1:100) | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | |- | ||
| + | | 10mM TrispH 8,5 | ||
| + | | 2,3 µl | ||
| + | | 2,3 µl | ||
| + | | 2,8 µl | ||
| + | | 2,8 µl | ||
| + | | 3,3 µl | ||
| + | | 3,3 µl | ||
| + | | 3 µl | ||
| + | | 3 µl | ||
| + | |- | ||
| + | |name of tube | ||
| + | | 14-F | ||
| + | | 14-R | ||
| + | | 21-F | ||
| + | | 21-R | ||
| + | | 13-F | ||
| + | | 13-R | ||
| + | | 34-F | ||
| + | | 34-R | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! | ||
| + | ! CMV for | ||
| + | ! CMV rev | ||
| + | ! BB018K5 for | ||
| + | ! BB018K5 rev | ||
| + | ! BB018K6 for | ||
| + | ! BB018K6 rev | ||
| + | ! BB021K17 | ||
| + | ! BB021K17 | ||
| + | |- | ||
| + | | template | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 3,8µl | ||
| + | | 3,8µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 3,8µl | ||
| + | | 3,8µl | ||
| + | |- | ||
| + | | primer (1:100) | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | | 3,2 µl | ||
| + | |- | ||
| + | | 10mM TrispH 8,5 | ||
| + | | 1,8 µl | ||
| + | | 1,8 µl | ||
| + | | 0 µl | ||
| + | | 0 µl | ||
| + | | 1,8 µl | ||
| + | | 1,8 µl | ||
| + | | 0 µl | ||
| + | | 0 µl | ||
| + | |- | ||
| + | |name of tube | ||
| + | | CMV-F | ||
| + | | CMV-R | ||
| + | | 185-F | ||
| + | | 185-R | ||
| + | | 186-F | ||
| + | | 186-R | ||
| + | | 2117-F | ||
| + | | 2117-R | ||
| + | |} | ||
| + | |||
| + | |||
| + | <font color="#009933">ligation BB020 BB022, Bb6 and BB5<sub>2</sub></font color> | ||
| + | |||
| + | -> Protocol [[Team:LMU-Munich/Notebook/Protocols/19 Transformation SSS|19 Transformation SSS]] | ||
| + | |||
| + | {| class="wikitable" border="1" | ||
| + | |- | ||
| + | ! | ||
| + | ! BB020 | ||
| + | ! BB022 | ||
| + | ! BB6 | ||
| + | ! BB5<sub>2</sub> | ||
| + | |- | ||
| + | | insert (5µl) | ||
| + | | 18+19 | ||
| + | | PCR 10 | ||
| + | | CMV | ||
| + | | PCR5<sub<2</sub> | ||
| + | |- | ||
| + | | insert (5µl) | ||
| + | | - | ||
| + | | - | ||
| + | | BB5 | ||
| + | | PCR6 | ||
| + | |- | ||
| + | | ccdB (1µl) | ||
| + | | c | ||
| + | | c | ||
| + | | c | ||
| + | | a | ||
| + | |- | ||
| + | | buffer | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | | 2µl | ||
| + | |- | ||
| + | | T4-ligase | ||
| + | | 0,5µl | ||
| + | | 0,5µl | ||
| + | | 0,5µl | ||
| + | | 0,5µl | ||
| + | |- | ||
| + | | H<sub>2</sub>O | ||
| + | | 11,5µl | ||
| + | | 11,5µl | ||
| + | | 6,5µl | ||
| + | | 6,5µl | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | <font color="#009933">PCR to test self-made taq</font color> | ||
| + | |||
| + | |||
| + | {| | ||
| + | | template eGFP | ||
| + | | 1µl | ||
| + | |- | ||
| + | | Primer 20,21 | ||
| + | | each 1µl | ||
| + | |- | ||
| + | | buffer (10x) | ||
| + | | 2µl | ||
| + | |- | ||
| + | | MgCl | ||
| + | | 2µl | ||
| + | |- | ||
| + | | dNTPs | ||
| + | | 0,2µl | ||
| + | |- | ||
| + | | taq | ||
| + | | 0,3µl to 0,7µl (0,1µl steps) | ||
| + | |- | ||
| + | | H<sub>2</sub>O | ||
| + | | fill to 20 µl | ||
| + | |- | ||
| + | |||
| + | |} | ||
| + | |||
| + | anealing temperature 58°C, elongation time 30'' | ||
| + | |||
{{:Team:LMU-Munich/Templates/Page Footer}} | {{:Team:LMU-Munich/Templates/Page Footer}} | ||
Latest revision as of 18:04, 27 October 2010
Short description of our work, our results and our supporters
Most genes and promoters were amplificated via PCR with overhang-primers with the BioBrick prefix or suffix. If the sequence contained a EcoR1-, Pst1-, Xba1-, Spe1- or Not1- restriction site, we used mutagenesis primers and fusioned both DNA parts by fusion PCR. All PCRs worked out, even the fusion PCRs.
The length of the PCR products were tested by agarose gel electrophoresis. We tried to sequence our PCR products, but obtained poor results and resorted to sequencing the plasmids.
In parallel, we made competent cells and multiplied ccdB (death gene)-vectors with different antibiotic resistances. All components were digested with the appropriate restriction enzymes. The samples were cleaned with a PCR clean up kit or dephosphorylated to reduce false ligations.
We ligated our constructs and several interim stages with the 3A-assembly according to our schedule. The ligations were transformed to E.coli DH5α strains and selected by antibiotics. Afterwards, some colonies were picked and we tested the insertion of the construct by colony PCR.
If the colony PCR resulted in bands of the right size, we extracted the plasmids from overnight cultures and sequenced the samples with forward and reverse BioBrick primers.
Unfortunately, not all BioBricks were cloned succesfully. However, we were able to produce 4 BioBricks, one of which represents a full construct while the other three are intermediates. The system wasn't completed on time, so we weren´t able to test them in eukarytic cell lines.
The protocols we used are listed here: Protocols
These Biobricks we submitted:
Sources, helpers and supporters:
Transforming competent cells
- eGFP Biobrick: BBa_I714891 SDY_eGFP (Kanamycin)
- TEV recogn N Degron SF3 = pDS7 (Ampicillin)
- TEV p14 recogn = 190-6 (Ampicillin)
-> Protocol: (3 Transformation)
- We added 2 µl DNA
- We plated out 200 µl
- CMV-Promoter Biobrick: BBa_J52034
-> Protocol:(4 Plasmid extraction from cells)
- Prepared overnight culture, measured concentration of DNA
-> Poor results -> thrown away
New Plasmid Extraction
- CMV-Promoter Biobrick: BBa_J52034
-> Protocol: (4 Plasmid extraction from cells)
- Plasmid concentration: 143ng/µl
- 3 ml LB-Media + 4 µl Kanamycin
- Inoculated with 1 colony of BBa_I714891 -> 37°C
- for 190-6 and pDS7: 10µl Ampicillin + 10 ml LB-Media + colony of plate
- for eGFP: 13,3 µl Kanamycin + 10 ml LB-Media + 1 colony of plate
-> mixed
- plus: EcoRI (10µg/µl): 0,5 µl resp. PstI (10µg/µl): 0,5 µl
- incubated at room temperature from 12:10 to 15:00, 1 hour at 37°C, 2 hours at 60°C
- frozen at -20°C
- 1 ml of "old" culture + 3 ml LB-Media + 4 µl Kanamycin -> 37°C
Plasmid Extraction of pDS7, eGFP, 190-6
-> Protocol: (4 Plasmid extraktion from cells)
- pDS7 (458ng/µl), eGFP (55ng/µl), 190-6 (193ng/µl)
Restriction digest of pDS7, eGFP, 190-6
- with EcoRI and PstI in buffer H (for testing DNA is correct)
-> Protocol: (5 Restriction digest)
- 10µg DNA: pDS7 (2µl), eGFP (15µl), 190-6 (10µl)
Plate colonies for plasmid extraction
- CMV (Kanamycin), eGFP (Kanamycin), pDS7 (Ampicillin), 190-6 (Ampicillin))
- PhiC31o plated on Ampicillin-Agar, stored at 37°C
50% Glycerol made
- for PhiC31o glycerol stock (produced later)
Inoculate CMV into LB medium with ampicillin
- CMV (BBa_J52034) from 10.8.2010 inoculated into LB medium with ampicillin, as falsly inoculated in Kanamycin
Agarosegelelectrophoresis with digestions
->Protocol (11 Agarose gel electrophoresis)
- Agarosegelelectrophoresis with the digestions (CMV, eGFP, pDS7, 190-6), 125V for 30 minutes and then for 20 minutes;
- expected DNA bands: 190-6 (4840bp, 1903bp), pDS7 (8027bp, 6bp), CMV (654 bp (Insert), 2079bp (Plasmid)), eGFP (720bp (Insert), 2750bp (Plasmid))
- Correct DNA bands for 190-6 (~4800bp, ~1900bp, ~6700bp (undigested plasmid)) and eGFP (~2000bp (Plasmid), ~750 bp (Insert)); CMV probably not digested (two bands; one probably normal, one supercoiled) and pDS7 not clear
Restriction digest from CMV and pDS7
-> Protocol (5 Restriction digest)
- Restriction digest from CMV (EcoR1, Pst1; 6µl DNA, buffer H) and pDS7 (EcoR1, Spe1; 2µl DNA, buffer B)
Agarosegelectrophoresis with digestions
->Protocol (11 Agarose gel electrophoresis)
- Agarosegelelectorphoresis for 30 minutes, 150V
- Expected DNA bands: CMV see above, pDS7 (3647bp, 3369bp, 1011bp, 6bp)
- false DNA bands CMV (~1200 bp, ~2000 bp) and pDS7 (~8000bp two bands, ~1100 bp); required to isolate a new colony for these two Plasmidextractions
Plated CMV on Ampicllin-Agar
- Plated the colony from CMV (BBa_J52034) for Plasmidextraction (Ampicillin), as falsly plated on Kanamycin
weekend
weekend
Planting colonies
- transfer 1 ml PhiC31o culture to new LB medium + Amp, 37°C
- pick up CMV and pDS7 colonies from plates and transfer to LB medium+Amp, 37°C
Plasmid Extraction of PhiC31o
->Protocol (4 Plasmid extraktion from cells)
- plasmid extraction of PhiC310
->27,5ng/µl DNA and second plasmid extraction of PhiC310 (i. o. to get more DNA); first eluation-step with first eluation-extraction
-> 60ng/µl DNA
Restriction digest
->Protocol (5 Restriction digest)
- restriction digest of PhiC310 with EcoR1 and Spe1
restriction digest in the thermo cycler (program "Verdau", see protocol)
Handling primers after arrival (1,2,3,4,5,6,11,12)
->Protocol (9 Handling primers)
PCR preparations
- 10mM dNTP mix made from 100 mM dATP, dGTP, dCTP, dTTP by taking 100µl of each and adding 600µl H 2 O
PCR 1 and 6
- PCR of the tet inducible CMV minimal promotor out of prevTRE (=PCR 1 with Primer 1 and 2) and SV40PA out of pcDNA3 (=PCR 6 with Primer 11 and 12)
->Protocol (10 PCR with Pfu)
Mixture:
Glycerolstock of PhiC31o
- Glycerolstock of the colony of PhiC31o for the plasmidextraction
Plate CMV and pDS7 colonies on Ampicillin-Agar
- colonies for plasmidextraction of CMV and pDS7 plated on Ampicillinplates
Plasmid Extraction of CMV and pDS7
- plasmidextraction of CMV (2,5ng/µl) and pDS7 (10ng/µl) the A260/A280 value was 1.333, which means that it was 90% Protein and only 10% DNA (should be 1,8); new plasmidextraction needed
new overnight cultures of CMV and pDS7 for a new plasmidextraction made
Agarose gel electrophoresis
-> Protocol (11 Agarose gel electrophoresis)
- Agarose gel electrophoresis of the restriction digest of PhiC31o and PCR 1 and 6
- the right bands found for PhiC31o (~2900,~2400,~250)
- the right band found for PCR1 (~450)
- no band found for PCR6; new electrophoresis needed with more DNA loaded
- new agarose gel electrophoresis from PCR6 with 5µl DNA instead of 3µl (image not yet shown)
- the right band found for PCR6 (~200)
New overnight cultures of CMV and pDS7
- the overnight colonies didn't grow; new colonies (CMV and pDS7) picked from plate and inoculated in LB Ampicillin
PCR purification of PCR 1 and 6
-> Protocol (12 Gel extraction or PCR Clean up)
- DNA concentration of the PCR 1 and 6 products measured: PCR1: 410ng/µl (A260/A280=1.253) PCR6: 568ng/µl (A260/A280=1.275)
- PCR Purification with Promega Kit
-> PCR1: 230ng/µl (A260/A280=1.769)
-> PCR6: 37.5ng/µl (A260/A280=1.667)
Plasmid Extraction of CMV and pDS7
-> Protocol (4 Plasmid extraction from cells)
- Plasmid extraction of CMV (97.5ng/µl; A260/A280=1.857) and pDS7 (212ng/µl; A260/A280=1.848)
Restriction digestion
-> Protocol (5 Restriction digestion)
- Restriction digestion of CMV (EcoR1 + Pst1; 10µl DNA, buffer H) and pDS7 (EcoR1 + Spe1; 5µl DNA, buffer B)
-> expected DNA bands: CMV: 2079bp (plasmid) + 654bp (Insert); pDS7: 7022bp + 1011bp
Agarose Gel electrophoresis of digested CMV and pDS7
-> Protocol (11 Agarorse gel electrophoresis)
-> right DNA bands for pDS7 (~7000bp, ~1000bp)
-> false DNA bands for CMV
- Starting PCR 2a and 2b (replication and mutagenesis of pDS7): 3 µl DNA and 50°C Annealing Temperatur (other same as 8-16-2010)
Agarose gel electrophoresis of PCR 2a and 2b
-> Protocol (11 Agarose gel electrophoresis)
(150V, 30min)
-> the right bands for PCR2a (~300bp) and PCR2b (~700bp)
- New agarose gel electrophoresis with all of the PCR product for gel extraction (150V, 30min)
Gel extraction of the DNA from PCR2a and PCR2b
-> Protocol (12 Gel extraction or PCR Clean up)
- DNA concentration measured; problem with nanodrop as too low concentration; lyophille used to reduce volume
- DNA concentration measured again: PCR2a: 70ng/µl A260/A280=1.647; PCR2b: 45ng/µl A260/A280=1.5
PCR 3 (joining PCR of 2a and 2b)
- PCR3 (the joining PCR of PCR2a and 2b; Joining of the TEVrecogn-N-Degron-SF3 part) done: 1.3 µl of PCR2a and 4.7 µl of PCR2b makes 300ng of a 1:1 solution of both to be joined DNA parts. Annealing temperature: 50°C
-> Protocol (10 PCR with Pfu)
Agarose gel electrophoresis of PCR3
left column: marker; rightmost column: PCR3
-> Protocol: 11 Agarose gel electrophoresis (150V, 30min)
-> expected band: ~1000bp
-> false band: ~500bp
- probable reason: mini photometre was influenced by gel extraction chemicals, therefore it measured false DNA concentrations and false template masses were calculated
-> New 2a and 2b PCR
New PCR (2a and 2b)
-> Protocol: 10 PCR with Pfu
(see 8-18-2010, but 35,5µl water)
weekend
weekend
Agarose gel electrophoresis of PCR 2b
-> Protocol: 11 Agarose gel electrophoresis
- expected band: 700bp
-> no band shown on gel -> new PCR 2b
PCR 2b
- start PCR 2b with PCR 2b from 8-13-10 as template ( 1:20 and 1:100 diluted; 1µl)
-> Protocol: 10 PCR with Pfu
- annealing temperature: 50°C; amount of water: 37,5µl
Agarose gel electrophoresis of PCR 2b 1:20 and 1:100
-> Protocol: 11 Agarose gel electrophoresis
- expected bands: each ~ 700bp
- false bands: ~ 200bp
-> new PCR with 2ng, 5ng, 10ng template pDS7
- pDS7 1:100 diluted(-> 2,1 ng/µl)
Mixture:
-> Protocol: 10 PCR with Pfu
PCR 2a gel extraction
- Quiagen kit (QuiaexII)
-> Protocol: 14 QIAEX II gel extraction
Start 3 CMV overnight cultures
agarose gel electrophoresis of PCR 2b
-> Protocol: 11 Agarose gel electrophoresis
- expected bands: right bands with 2ng and 5ng template (~700bp), no band with 10ng template
CMV plasmid extraction
-> Protocol: 4 Plasmid extraction from cells
Plasmid extractionof 3 different overnight cultures.
- results:
CMV restriction digestion
-> Protocol: 5 Restriction digestion
- CMV restriction digest: EcoRI, PstI, buffer H
PCR 2b gel extraction
- PCR2b was gel extracted (with Qiagen gel extraction kit), 17.5 ng/µl a260/A280= 1.750
-> Protocol: 14 QIAEX II gel extraction
PCR 3 (fusion of 2a and 2b)
- PCR3: conducted again at 52°C annealing temperature
-> Protocol: 10 PCR with Pfu
agarose gel electrophoresis of CMV digestion
- agarose gel electrophoresis (150V, 25 min) of the CMV digestion
-> bands are wrong again ( ~ 1200bp, 2000bp)
Agarose gel electrophorese of PCR 3
-> Protocol: 11 Agarose gel electrophoresis
- expected band: ~1000bp
- false band: ~400bp
Plasmid extraction of ccdB tet and ccdB strep
Plasmid extraction of pSB1C3 with BBa_P1010
-> Protocol: 4 Plasmid extraction from cells
- results:
- plate ccdB with ampicilline, chloramphenicol, tetracycline resistance on LB agar with appropiate antibiotic.
- Overnight culture of ccdB with kanamycine resistance in LB medium with kanamycine
PCR 7a, 7b, 9, 10
->Protocol: 10 PCR with Pfu
Standard PCR; annealing temperature: 60°C
Agarose gelelectrophoresis of PCR 7a, 7b, 9, 10
->Protocol: 11 Agarose gel electrophoresis
- 150V, 25min
Plasmid extraction of ccdB kan
-> Protocol: 4 Plasmid extraction from cells
-result: concentration: 25ng/µl; A260/A280= 2,0
New PCR 7a, 7b, 9, 10
Mixture
Program:
gradient PCR, 42-69°C annealing temp.
->Protocol: 10 PCR with Pfu
Overnight culture of ccdB amp, tet, cam
Inoculate one colony each in 5ml medium with approptraite antibiotic.
Agarose gel electrophoresis of PCR 7a, 7b, 9, 10
->Protocol: 11 Agarose gel electrophoresis
150V, 25min, 75mA
from left to right: 7a, 7b, 9, 10, Marker
Plasmid extraktion of ccdB amp, tet, cam
->Protocol: 4 Plasmid extraction from cells
results:
Mixture:
- 2ng template: see 26-8-10
- 4ng template: see 26-8-10, but 2µl template and 35,25µl water
-> Protocol: 10 PCR with Pfu
-> Protocol: 5 Restriction digestion
- only 90min 37°C incubation
- EcoRI, PstI, Buffer H
Agarose gelelectrophoresis of PCR 7a, 9, ccdB restriction digestion
150v, 25min, 75mA
-> Protocol: 11 Agarose gel electrophoresis
results:
- PCR7a, 9: false band at 200bp
- ccdB: each digestion leads to a right band with ~ 650bp
weekend
weekend
New PCR 7a and 9
PCR 7: Annealing Temperature 60°C - 25 x 1 min Annealing time and 5x 1,30 min Annealing time
PCR 9: Annealing Temperature 55°C - 25 x 1 min Annealing time and 5x 1,30 min Annealing time
->Protocol: 14 QIAEX II gel extraction
results:
-> Protocol: 11 Agarose gel electrophoresis
150V, 25min
- results:
- 7a: no band shown
- 9: false band (~200bp)
New PCR 3
-> Protocol: 10 PCR with Pfu
- new method: standard PCR without primers (10 cycles, 56°C annealing temp.)
- then add 2,5µl of primer 3 and 6
- 30 cycles standard PCR (54°C annealing temp.)
Agarose gel electrophoresis of PCR 4a, PCR4b, PCR3(Pfu), PCR3(Phusion)
-> Protocol: 11 Agarose gel electrophoresis
150V, 25min
- results:
- PCR4a(2.5ng template), PCR4a(5ng template),PCR4b(2.5ng template), PCR4b(5ng template), PCR3(Pfu): no band shown
- PCR3 (Phusion): right band (~1000bp)
New PCR PCR4a, PCR4b, PCR7a, PCR9
-> Protocol: 10 PCR with Pfu
PCR mixture for PCR4a, PCR4b
Standard PCR program with annealing temperature PCR4a: 51.1°C, PCR4b: 48.5°C.
-> Protocol: 15 PCR with Phusion
PCR mixture for PCR7a, PCR9
PCR program: Phu62
Agarose gel electrophoresis of PCR 3, PCR7a, PCR9 for gel extraction
-> Protocol: 11 Agarose gel electrophoresis
120V, 30min
- results:
- PCR3; right band (~1000bp) and side-product
- PCR7a: no band
- PCR9: right band (~800bp) and side-product
Gel extraction of the DNA from PCR3 and PCR9
-> Protocol (12 Gel extraction or PCR Clean up)
results:
PCR 9: 22,5ng/µl; A260/A280=1,8
PCR 3: 22,5ng/µl; A260/A280=2,25
New PCR 4a, 4b, 7a with DreamTaq
-> Protocol: 16 PCR with DreamTaq
PCR mixture for PCR7a
Primers for PCR 7a: 13,14
Annealing temp: 60°C
PCR mixture for PCR4a,4b
PCR 4a
Primers for PCR 4a: 7,8
Primers for PCR 4b: 9,10
Annealing temp: 50°C
PCR program:
Agarose gel electrophoresis of PCR4a, PCR4b, PCR7 (DreamTaq)
-> Protocol: 11 Agarose gel electrophoresis
150V, 25min
results: no product
New PCR 4a, 4b with DreamTaq, Pfu, with concentration gradient and touch-down PCR
-> Protocol: 16 PCR with DreamTaq; 10 PCR with Pfu
PCR mixture for DreamTaq
PCR mixture for Pfu
Primers for PCR 4a: 7,8; PCR 4b: 9,10
-> Protocol: Thermal cycler program: Touch down
Agarose gel electrophoresis of PCR4a, PCR4b
-> Protocol: 11 Agarose gel electrophoresis
150V, 25min
from left to right: 4a: P1, P2, P3, D1, D2, D3; 4b: P1, P2, P3, D1, D2, D3
key:
"P"= PCR with Pfu
"D"= PCR with DreamTaq
"1"= low template concentration
"2"= middle template concentration
"3"= high template concentration
expected bands:
Agarose gel electrophorese of PCR 4a P2, 4b P2
-> Protocol: 11 Agarose gel electrophoresis
- 120V, 45min, 1,5% Agarose gel
Agarose gel electrophoresis of (from left to right) PCR4aP2, Marker and PCR4bP2
PCR Agarose gel extraction
-> Protocol: 14 QIAEX II gel extraction
results:
template: 190-6, Primer 13,14
Mixture with Pfu
PCR mixture with Phusion
-> Protocol: 15 PCR with Phusion
New PCR 4a, 4b with Pfu
-> Protocol: 10 PCR with Pfu
-> Protocol: Thermal cycler program: Touch down
Mixture see 9-1-10, twice 4a and 4b
Agarose gel electrophorese of PCR 4a, 4b, 7a, gel extracted 4a and 4b
-> Protocol: 11 Agarose gel electrophoresis
- 25min, 150V
from left to right: 4a*, 4a, 4b*, 4b, 7a Phusion, 7a Pfu, Ladder
-> result: 4b, 4b*: right bands (~330bp)
-remain: false bands/no band
from left to right: ladder, 4 columns pathway, 4a gelextr., 4b gelextr.
-> results: slight right bands for 4a and 4b, no "primer clouds" anymore.
Overlapping PCR 5 with Pfu and Phusion
template: 4a, 4b Primer 7,10
Mixture with Pfu
-> Protocol: 10 PCR with Pfu
PCR program: standard PCR program for Pfu, Annealing temperature: 54°C
PCR mixture with Phusion
-> Protocol: 15 PCR with Phusion
PCR program: standard PCR program for Phusion, Annealing temperature: 58°C
New PCR7a with Pfu
template: 190-6; Primer: 13,14
Mixture with Pfu
-> Protocol: 10 PCR with Pfu
PCR program: standard PCR program for Pfu, Gradient: 54.4°C, 57.8°C, 61.4°C, 65.0°C
-> results:
-7a: only "primer cluods"
-5 Pfu: no defined product (slurred?)
-5 Phusion: "primer clouds"
weekend
weekend
charges for sequencing
every charge 7µl
template: 4a, 4b; 15ng
Primer 7,10
Mixture
-> Protocol: 10 PCR with Pfu
PCR program: standard PCR program for Pfu, Annealing temperature: 46.1°C,48.5°C, 51.1°C
-> results: "primer clouds"
-> results: "primer clouds"
every charge 7µl
Agarose gel electrophorese of PCR gel extractions
150V, 25min
-> Protocol: 11 Agarose gel electrophoresis
-result:
from left to right: PCR 1,2a,2b,3,4a,/,7b,9,10,/,ladder,pathway
new PCR for PCR6
PCR mixture for PCR6
-> Protocol: Thermal cycler program: Touch down, 62°C-52°C, 30 cycles by 55°C
Restriction digestion of eGFP, PCR6, ccdBamp
as prepatation for ligation
-> Protocol 5 Restriction digestion
1:30h 37°C, 20min 80°C
Agarose gel electrophoresis of PCR6
-> Protocol: 11 Agarose gel electrophoresis
25min, 150V
Concentration of DNA in Ligation 1:
-> Protocol 18 competent cells2
The incubation time for the cells is here 1 hour.
Agarose gel electrophoresis of PCR 1, 3, 4a, 5, 7a, 9
Agarose gel electrophoresis of PCR 1, 3, 4a, 5, 7a, 9
-> protocol 11 Agarose gel electrophoresis
results: "primer-clouds", PCR 9: no band
-> Protocol 4 Plasmid extraction from cells
Eluated with H2O instead of the Eluation Buffer.
New PCR 1, 3, 4a, 5, 7a, 9, 10 with phusion
+ in each assay:
dNTP mix: 1µl, 5x Phusion buffer: 10µl, DMSO:1.5µl, Phusion:0.5µl
sum: 50µl
Program: standard Phusion PCR, 29 cycles; annealing temperatures: see above
-> Protocol: 15 PCR with Phusion
[from left to right: PCR 1, 3, 4a, 5, 7a, 9, 10, Ladder]
PCR3, 9, 10 with right bands.
New PCR 1, 4a, 4b, 5, 7a with phusion
+ each assay with dNTP-mix (1µl), 5xBuffer (10µl, DMSO (1.5µl), Phusion (0.5µl)
-> sum: 50µl
-> Protocol Touch down 59 with phusion, 30 cycles with gradient appropriate for the annealing temperatures above.
Agarose gel electrophoresis of PCR 1, 4a, 4b, 5, 7a
-> Protocol: 11 Agarose gel electrophoresis
[From left to right: Ladder, 1, 4a, 4b, 5, 7a]
Only 4a has been amplified successfully.
from left to right: PCR 3, band ~700bp, PCR 4a, band ~300bp, PCR 5, bands ~550bp (5*), ~650bp (5), PCR 9, band ~800bp, PCR 10, band ~1900bp
results:
-> Protocol 12 Gel extraction or PCR Clean up (Promega kit)
weekend
weekend
charges for sequencing (retry of 6.9.)
every charge 7µl
template: p190-6
Primer 13,14
Mixture
PCR program: touchdown PCR with Taq
-> Protocol: 21 PCR with Taq Mastermix
overnight culture inoculated of
with 2ng and 4 ng template
total: 100µl each, divided into 5 charges
program:
-> Protocol:20 PCR with Phusion Hot Start
Agarose gelelectrophoresis of PCR 7a
-> Protocol: 11 Agarose gel electrophoresis
bands 1 to 5: 2ng of template DNA
bands 6 to 10: 4ng of template DNA
-> no bands
new PCR 7a and PCR 8 (without mutation)
with mastermix, without DMSO
7a diluated and undiluated: divided into 3 charges
7a diluated: 1,2,3; 7a undiluated: 4,5,6
program:
Plasmid extraction of ccdB (amp) and ccdB (cam)
-> Protocol: 4 Plasmid extraction from cells
results:
Agarose gelelectrophoresis of PCR 7a diluted &7a undiluted & "8"(without mutation)
-> Protocol: 11 Agarose gel electrophoresis
from left to right: 7a: 1:100 diluted: 48°C, 52°C, 56.1°C, undiluted: 48°C, 52°C, 56.1°C, 8 without Mutation
weak right band (with fuzz)for 7a undiluated with best result for 56°C
-> new PCR 7a and "8"
with mastermix Taq
-> Protocol: 21 PCR with Taq Mastermix
template: 190-6, 1:10 and 1:5
temp: 56°C and 58°C
2 charges each: one for 56°C and one for 58°C annealing temperature
EcoR1 + Pst1 with Buffer H; 50ng DNA
-> Protocol: 5 Restriction digestion
Agarose gelelectrophoresis of PCR 7a 1-4 & "8" 1-4(without mutation)
-> Protocol: 11 Agarose gel electrophoresis
-> bad results
purification of restriction digestion
-> Protocol: 12 Gel extraction or PCR Clean up (Promega kit)
Ligation with pSB1C3 (2072bp)
plus 1 µl T4 ligase in each charge
-> Protocol: 22 Ligation
with mastermix Taq 56°C
sum: 20µl each
pcr-program:
-> Protocol: 21 PCR with Taq Mastermix
Agarose gelelectrophoresis of PCR 7a-1, PCR 7a-2, PCR 8-1, 8-2 and of all three PCR 6 we ever made (in order to control whether we have extracted the right fragment (237) or just the primerdimers)
-> Protocol: 11 Agarose gel electrophoresis
From left to right: 62, 61, 6, Ladder, 7a-1, 7a-2, 8-2, 8-1
transformation of the ligations 1,3,51, 52, 6, 9, 10
-> Protocol:3 Transformation
->plated on plates with chloramphenicol
again PCR 1,3,5,6,9,10 in order to increase the profit (Ausbeute) for ligation
pcr-program:
-> Protocol: 21 PCR with Taq Mastermix
Analysis of the transformation from yesterday
Colonies on plates when 100µl or pellet plated:
again: transformation of ligation 1
->Protocol: 3 Transformation
Agarose gelelectrophoresis of yesterday's PCR 1,3,5,6,9,10(without mutation)
-> Protocol: 11 Agarose gel electrophoresis
From left to right: PCR1, PCR3, PCR5, Ladder, PCR6, PCR9, PCR10
-> right band for PCR 3,5,6,9,10; no band for PCR 1
From left to right: Ladder, PCR3, PCR5, PCR9, PCR10
Bands took: 32: Band short over 1000bp, 31: Band short under 1000bp (probably 32 right), 5 the highest band, 9 upper band, 10 upper band
new PCR 1,5,6
-> Protocol:
pcr-program: standard Pfu 10 PCR with Pfu
-> Protocol:5 Restriction digestion
Colony PCR of Ligations 3, 51, 52, 6, 9, 10 and PCR of CMV1 (to test if right)
10 1-4, 9 1-4, 6 1-4, 51 1-4, 52 1-4, 3 1-4 Colonies picked and put in following Mix:
CMV1:
-> Protocol: 21 PCR with Taq Mastermix
Agarose Gel electrophoresis of 3, 51, 52, 6, 9, 10 and CMV1 (to test if right)
-> Protocol: 11 Agarose gel electrophoresis
Above: From left to right: 3.1, 3.2, 3.3, 3.4, 51.1, 51.2, 51.3, 51.4, Ladder, 52.1, 52.2, 52.3, 52.4
Below: From left to right: 6.1, 6.2, 6.3, 6.4, 9.1, 9.2, 9.3, 9.4, Ladder, 10.1, 10.2, 10.3, 10.4, CMV1
-> right bands for 6.3 and 6.4, CMV1 ~1200bp (we think that this is right, as Biobrick sequenzing information indicates that it isn't 654bp but about 1200bp)
-> Protocol: 12 Gel extraction or PCR Clean up (Promega kit)
weekend
weekend
Dephosphorylation of linearized ccdB amp and ccdB cam
Add 1µl TSAP to digested vectors.
Incutation: 37°C 15min; Inhibition: 74°C 15min
Ligation and 3A-Assemblies
Jump-or-Die Ligations: 1a, 1b (51),1b (52), 2, 3
Cut'N'Survive Ligations: 1a, 2b,
Biobricks for both Systems: CMV, PCR1 (tet-on-promoter)
Mixtures:
- 3A Assemblies (everything except CMV, PCR1):
Inserts: each 8µl; ccdB vector: 1µl; T4 Ligase: 0.5µl; T4 Ligase Buffer: 2µl; H2O: 1µl
- Biobricks:
Insert: 10µl; ccdB vector: 1µl; T4 Ligase: 0.5µl; T4 Ligase Buffer: 2µl; H2O: 7µl
Incubation: 2:30h 16°C; Inhibition: 10 min 65°C
->Protocols: 22 Ligation, 13 3A Method for Biobrick assembly
Colony PCR of PCR product ligations with pSB1C3
Mixture:
PCR Mastermix:130µl ; PrimerF:19,5µl ; PrimerR:19,5µl ; H2O:91µl
-> Protocol: 21 PCR with Taq Mastermix
Agarose Gel Electrophoresis of Colony PCRs
-> Protocol: 11 Agarose gel electrophoresis
above from left to right: pathway/ladder/3.5/3.6/3.7/3.8/51.5/51.6/51.7/51.8/52.5/52.6/52.7/52.8
below from left to right: ladder/6.5/6.6/6.7/6.8/9.5/9.6/9.7/9.8/10.5/10.6/10.7/10.8
Agarose Gel Electrophoresis of PCRs 1, 5, 6, and Colony PCR 3.8, 51.7, 52.6, 9.7, 10.5, 10.6
-> Bad results, probably too much cells in PCR mix
-> Protocol: 11 Agarose gel electrophoresis
Colony PCR of 3.8, 51.7, 52.6, 9.7, 10.5, 10.6 again and of the 3A Ligations of yesterday as well as the Ligation of PCR1 and CMV with pSB1C3
PCR Master Mix: 60µl
Primer F: 9µl
Primer R: 9µl
H2O: 42µl
->22charges à 5µl
-> Protocol:21 PCR with Taq Mastermix
sum: 50µl
same with Primer 13l (long)
->Protocol: 21 PCR with Taq Mastermix
Agarose Gel Electrophoresis of Colony PCRs and PCR 7a
from left to right:7ak(short primer), 7al(long primer), JD.1a.1, JD.1a.2,JD.1b51.1, JD.1b52.2, JD.1b52.1, JD.1b52.2, JD.2.1, JD.2.2, JD.31.1, JD.31.2, CS.1a.1, CS.1a.2
no bands shown: CS.2b.1, CS.2b.2, PCR1.1, PCR1.CMV,3.8, 51.7, 52.6, 9.7, 10.5, 10.6
-> Protocol: 11 Agarose gel electrophoresis
Inoculate Colonies
inoculated in 4ml LB-medium with appropriate antibiotic
Plated residual transformated E. colis (9-20-10) (where we had few colonies on plates)
Colony PCRs
for 22*5µl charges:
-> Protocol: 21 PCR with Taq Mastermix
Annealing temperature: 53°C
charges 7ak and 7al had not been labeled, so they were renamed.
results:
7a1: 135ng/µl; A260/A280=1.7
7a2: 133ng/µl; A260/A280=1.7
-> Protocol: 20 PCR with Phusion Hot Start
Program: annealing temperature: 63°C, elongation time: 40sec
Agarose gel electrophoresis of Colony PCRs and PCR8
new ladder: fermentas FastRuler (TM) Middle Range DNA Ladder http://www.fermentas.com/en/products/all/dna-electrophoresis/fastruler-dna-ladders/sm111-fastruler-middle
some results:
PCR 8 (lanes 23, 24): no band
Colony PCRs CS.2b.10, PCR1cam.3, PCR1cam.4, CMVcam.4, CMVcam.4, JD.1b51.10 (lanes 7,8,9,10,11,14): show right bands
->Protocol: 4 Plasmid extraction from cells
concentrations (ng/µl - A260/280):
new PCR 7b in order to get better results
overnight-cultures
inoculated of: CS2bK10=BB11; PCP1K3+4=BB1; CMV-C-K3+4; JD1b51K10=BB2; pSB1C3
Restriction digestion of BB5 for the 3A Method
sum: 20µl
-> Protocol:5 Restriction digestion
Plasmid extraction of overnight cultures BB2, BB11, CMV in cam and pSB1C3
->Protocol: 4 Plasmid extraction from cells
concentrations (ng/µl - A260/280):
The pSB1C3 colony was pink (produced a red pigment or something like that, so we make a new transformation in order to extract only pSB1C3 and not some other vectors.
Agarose gelelectrophoresis of PCR 7b and pathway restriction digestions
-> Protocol: 11 Agarose gel electrophoresis
From left to right: PCR7b, pathway (3x),ladder (5000,2000,850,400,100bp)
result: PCR 7b shows right band with ~400bp
PCR clean up of PCR 7b
-> Protocol: 12 Gel extraction or PCR Clean up (Promega kit)
result: 90ng/µl; A260/A280= 1,895
Restriction digestion of BB2 and BB11
sum: 20µl
sum: 20µl
-> Protocol 5 Restriction digestion
-> Protocol: 22 Ligation
PCR 8 with 7b(new) plus 7a-2 with Phusion Hot Start II; Primer 16 plus 13k and 13l
Overnight culture of PCR1 in cam- Vector
new inoculation of PCR1 in cam, Colony 3 in 5ml LB+cam
Transformation of pSB1C3, BB1, BB6
New transformation of pSB1C3 from Spring distribution because the old colony was pink.
Transformation of BB1 and BB6.
-> Protocol: 19 Transformation (Sina Science Services)
Agarose gelelectrophoresis of PCR8 (long and short primer 13)
PCR8k=short primer 13, PCR8l=long primer 13
-> Protocol: 11 Agarose gel electrophoresis
result: PCR 8k and 8l shows right band with ~1200bp
PCR purification of PCR 8
-> Protocol: 12 Gel extraction or PCR Clean up (Promega kit)
result: ~70ng/µl; A260/A280=1.75
New "over-day" culture of pSB1C3
Inoculate one culture from the ligation yesterday in 4ml LB+cam
Ligation of BB72
Mixture:
Inserts: each 5µl; ccdB vector: 1µl; T4 Ligase: 0.5µl; T4 Ligase Buffer: 2µl; H2O: 7µl
Incubation: 5h 18°C; Inhibition: 20 min 65°C
->Protocols: 22 Ligation, 13 3A Method for Biobrick assembly
Colony PCRs of BB1 and BB6
2 Colonys were picked from each plate -> 4 charges
Mixture for 4 charges with 5µl:
+ Colony
-> Protocol: 21 PCR with Taq Mastermix
Annealing temperature: 53°C, standard protocol
Plasmid extraction of new PCR1 in cam vector
-> Protocol: 4 Plasmid extraction from cells
results:
-> Protocol: 4 Plasmid extraction from cells
results:
weekend
weekend
Colony PCR of BB72, BB6 and BB1 plus a negativ charge as reference
-> Protocol 24 Colony PCR
-> Protocol 22 Ligation
-> Protocol 22 Ligation
Restriction digestion of PCR32, 8l, 10 and Primer 18+19 with E+P
-> Protocol 5 Restriction digestion
Plating BB017, BB021, BB018, BB001, BB72, BB10
Colony PCR BB1, BB6, BB72 with DreamTaq (each 2 colonies)
-> Protocol: 16 PCR with DreamTaq
PCR mixture for PCR7a
Primers for colony PCR: BBseqF, BBseqR
Annealing temperature: 53°C
Ligation of BB2, BB019, BB020, BB016, BB022
plates BB72, BB017, BB10, BB021, BB001, BB018: nothing grown -> thrown into trash
colony-PCR of BB2/8, BB019, BB016, BB020, BB022 with Taq
->protocol: PCR with Taq
template: pick colony
sum: 10µl; we made 100µl ->10 charges (2 colonies of each plate)
tube numbers:
PCR-program: taq53
agarose-gelelektrophoresis -> no bands visible
new plates made of BB1, BB4, BB6, BB7
Ligations BB72, BB017 (mit PCR51), BB10, BB021, BB001, BB018
restriction degestions
of PCR6, PCR52, PCR32, PCR1, ccdB(amp), ccdB(cam)
->protocol: 5 Restriction digestion
->protocol: PCR with Taq
Mastermix:
->10µl for each charge
template: pick colony; BB11: 2ng
tube numbers:
PCR program:TA = 53°C; elongation time=1min
Colony PCR of BB2/BB8, BB6 K7&K8 and test PCR of BB11 with new dilutions and primers
-> Protocol 24 Colony PCR
PCR purification of the digestions of 6I, 6II, 52, 32, 1
-> Protocol 12 Gel extraction or PCR Clean up (Promega kit)
Transformation of BB018, BB72, BB001, BB017, BB10 and BB021
-> Protocol 19 Transformation SSS
Plasmidextraction of the following overnight cultures: BB020(K1), BB018(K5), BB6(K5), BB022(K1), BB016(K1), BB019(K1), BB2(K1) and BB018(K3)
-> Protocol 4 Plasmid extraction from cells
new test PCR of BB5 and BB11 with 20µl and 50µl volume
Master Mix
template: BB5/50µl: 5ng, 20µl: 1.5ng; BB11/20µl: 3ng
->protocol: PCR with Taq
from left to right: ladder, BB5/50µl, BB5/20µl, BB11/20µl. BB5/50µl and BB11/20µl show the right band.
weekend
weekend
sequencing
cycle, clean and run
Colony PCRs of BB6, BB4, BB021, BB022, CMV©, BB001, BB016, BB1, BB252, BB72, BB017, BB018, BB019, BB020
agarose gel electrophoresis
schedule of the agarose gel electrophoresis:
Gelbilder und Auswertung einfügen
overnight cultures of BB6K3, BB4K10, CMV©K4, BB001K5, BB001K9, BB016K11, BB1K8, BB252K3, BB72, BB017K14, BB018K10, BB019K6
Plasmidextraction of the following overnight cultures: BB6(K13), BB4(K13), CMVc (K4), BB001(K5), BB001(K9), BB016(K11), BB1(K8), BB2(K3), BB7(K10), BB017(K14), BB018(K10) and BB019(K6)
-> Protocol 4 Plasmid extraction from cells
colony PCR of BB021, BB022 with Taq (53°C)
plasmide extractions from 10-06-2010 were put in a lyophilizer and redissolved in less water (10-20 µl)
Plasmid extraction BB018K5, BB018K6, BB021K17 and BB021K24
concentration of these samples to be measured on Monday
-> Protocol 4 Plasmid extraction from cells
weekend
Plasmid extraction BB018K5, BB018K6, BB021K24 and BB021K17 from 10-09-2010
foto einfügen!
Sequencing 8,9 and 10 (cycle, clean and run)
-> Protocol 19 Transformation SSS
PCR to test self-made taq
anealing temperature 58°C, elongation time 30


![]()
![]()







![]()
![]()

![]()
ApoControl Notebook
What we did
The creation of certain constructs was necessary for our two systems for cell selection by means of apoptosis: “Cut’N’Survive” and “Jump-Or-Die”. We searched for sources of the DNA sequences we needed and found several supporters which are listed below.
Contents>
8-10-2010
Plasmid Isolation
8-11-2010
Prepared overnight culture of eGFP BBa_I714891
Prepared overnight culture of 190-6 and pDS7 and eGFP (BBa_I714891) in falcons
Restriction digestion of CMV-Promoter BBa_J52034 with EcoRI and PstI
H2Oddest, sterile
10,3 µl
RE10 + Buffer H
2,0 µl
acetylated BSA (18ng/µl)
0,2 µl
DNA (0,143µg/µl)
6,0 µl
Prepare new/fresh overnight culture of CMV-Promoter Biobrick: BBa_J52034
8-12-2010
8-13-2010
8-14-2010
8-15-2010
8-16-2010
H2Oddest, sterile
0 µl
Buffer B
2,0 µl
BSA (1:10)
2 µl
DNA (0,06µg/µl)
15,0 µl
EcoR1
0,5 µl
Spe1
0,5µl
pTRERev (0,15µg/µl)
pcDNA3 (0,6 µg/µl)
Primer
2*2,5µl (P1+P2)
" (P11+P12)
300ng template
0,5µl
2µl
10x Buffer Pfu
5µl
"
dNTP Mix
1µl
"
Pfu Polymerase (3u/µl)
0,5µl
"
H2O
40,5µl
39µl
summ
52,5µl
52,5µl
Programme:
Denaturation
95°C
2min
30 times:
Denaturation
95°C
1min
Annealing
45°C
30sec
Extension
73°C
2min
Final Extension
73°C
5min
Soak (end)
12°C
infinite
bacterial culture
800µl
Glycerol (50%)
500µl
8-17-2010


Agarose gel electrophoresis of (from left to right) PhiC31o, PCR1 and PCR6
Agarose gel electrophoresis of (from left to right) PhiC31o, PCR1 and PCR6 which shows that PCR1 is between 250 and 500 bp
8-18-2010
8-19-2010
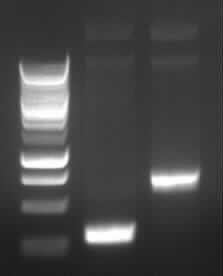
Agarose gel electrophoresis of (from left to right) PCR2a and PCR2b
8-20-2010
8-21-2010
8-22-2010
8-23-2010
PCR 2b with 2ng, 5ng, 10ng template pDS7
2ng
5ng
10ng
Primer
2*2,5µl (P5+P6)
2*2,5µl (P5+P6)
2*2,5µl (P5+P6)
10x Buffer Pfu
5µl
5µl
5µl
dNTP Mix
1µl
1µl
1µl
template
pDS7 (dil.)
1µl
2,5µl
5µl
Pfu Polymerase (3u/µl)
0,5µl
0,5µl
0,5µl
DMSO
1,25µl
1,25µl
1,25µl
H2O
33,25µl
30,25µl
25,25µl
sum
8-24-2010
PCR2a
0.9 µl
PCR2b
0.6 µl
primer3
2.5 µl
primer6
2.5 µl
dNTPs
1 µl
Pfu
0.5 µl
10xbuffer
5 µl
H2O
37 µl
8-25-2010
ccdB tet:
50ng/µl;
A260/A280= 1,818
Plate ccdB amp, cam, tet
Overnight culture of ccdB kan
PCR nr.
template
concentration
dilution
primer
7a
190-6
~200ng/µl
1:100
13,14
7b
190-6
~200ng/µl
1:100
15,16
9
eGFP
55ng/µl
1:25
20,21
10
PhiC31o
20ng/µl
1:10
22,23
Mixture
template (~2ng)
1µl
Pfu
0,5µl
Primer *2
2,5µl *2
10x buffer
5µl
dNTP Mix
1µl
H2O
37,5µl
sum
50µl
8-26-2010
PCR nr.
expected bands
result
7a
850bp
no band
7b
402bp
false band (200bp)
9
808bp
no band
10
1888bp
no band
template (~2ng)
1µl
Pfu
0,5µl
Primer *2
2,5µl *2
10x buffer
5µl
dNTP Mix
1µl
DMSO
1,25µl
H2O
36,25µl
sum
50µl
8-27-2010
PCR nr.
expected bands
result
7a
850bp
no band
7b
402bp
right band (~400bp)+ false band (~150bp)
9
808bp
false band (~200bp)
10
1888bp
right band (~1900bp)+false band (~500bp)
Plasmid
concentration
A260/A280
ccdB amp
57,5 ng/µl
1,917
ccdB cam
70,0 ng/µl
1,867
ccdB tet
50,0 ng/µl
1,818
New PCR 7a, 9
Restriction digestion of ccdB amp, kan, cam, tet
template
volume
mass
ccdB amp
16µl
930ng
ccdB cam
14,3µl
1µg
ccdB tet
16µl
800ng
ccdB kan
16µl
400ng
8-28-2010
8-29-2010
8-30-2010
Mixture
template (~4ng)
2µl
Pfu
0,5µl
Primer *2
2,5µl *2
10x buffer
5µl
dNTP Mix
1µl
DMSO
1,25µl
H2O
35,25µl
sum
50µl
-> Protocol: 10 PCR with Pfu
PCR program
Gel extraction of PCR 7b, 10
PCR nr.
concentration
A260/A280
7b
10 ng/µl
2,0
10
17,5 ng/µl
1,4
Agarose gel electrophoresis of new PCR 7a, 9
PCR2a
0.9 µl
PCR2b
0.6 µl
dNTPs
1 µl
Pfu
0.5 µl
10xbuffer
5 µl
DMSO
1,25µl
H2O
36,75 µl
sum
45µl
8-31-2010
template
37.25 µl (200ng)
dNTPs
1 µl
Pfu
0.5 µl
10xbuffer
5 µl
DMSO
1,25µl
sum
50µl
template
4 µl (8ng)
dNTPs
1 µl
Phusion
0.5 µl
5xbuffer
10 µl
DMSO
1,25µl
H2O
28.25 µl
sum
50µl
98°C
1 min
98°C
10 sec
62°C
20 sec
73°C
30 sec
return to step 2 for 29 cycles
73°C
10 min
12°C
forever
template
5 µl (10ng)
dNTPs
5 µl
DreamTaq
0.33 µl
10xbuffer
5 µl
DMSO
1,25µl
H2O
28.5 µl
sum
50µl
template
36,5 µl (180ng HeLa cDNA)
dNTPs
5 µl
DreamTaq
0.33 µl
10xbuffer
5 µl
DMSO
1,25µl
sum
50µl
95°C
1 min
95°C
30 sec
50/60°C
30 sec
72°C
1 min (1kb/min)
return to step 2 for 29 cycles
72°C
10 min
12°C
forever
9-01-2010
Concentration
Low
Middle
High
template
5 µl (1:1000)
31.75µl (1:1000)
2µl (1:10)
dNTPs
5 µl
DreamTaq
0.33 µl
10xbuffer
5 µl
DMSO
1,25µl
H2O
26.75 µl
0
29.75
sum
50µl
Concentration
Low
Middle
High
template
5 µl (1:1000)
37.25µl (1:1000)
2µl (1:10)
dNTPs
1 µl
Pfu
0.5 µl
10xbuffer
5 µl
DMSO
1,25µl
H2O
32.25 µl
0
35.25 µl
sum
50µl
9-02-2010
- Cut out bands at ~~ 350bp and extract
PCR nr.
concentration
A260/A280
4aP2
12,5 ng/µl
1,67
4bP2
72,5 ng/µl
1,53
New PCR 7a with Pfu and Phusion
template (~2ng)
1µl
Pfu
0,5µl
Primer *2
2,5µl *2
10x buffer
5µl
dNTP Mix
1µl
DMSO
1,25µl
H2O
36,25µl
sum
50µl
-> Protocol: 10 PCR with Pfu
template (~2ng)
1,0 µl
dNTPs
1 µl
Phusion
0.5 µl
5xbuffer
10 µl
DMSO
1,25µl
H2O
31,25µl
sum
50µl
9-03-2010
PCR4a (~15ng)
1.2µl
PCR4b (~15ng)
2µl (1:10)
Pfu
0.5µl
Primer *2
2.5µl *2
10x buffer
5µl
dNTP Mix
1µl
DMSO
1,25µl
H2O
34.05µl
sum
50µl
PCR4a (~15ng)
1.2µl
PCR4b (~15ng)
2µl (1:10)
dNTPs
1 µl
Phusion
0.5 µl
5xbuffer
10 µl
DMSO
1,25µl
H2O
29.05µl
sum
50µl
template (~2ng)
1µl
Pfu
0.5µl
Primer *2
2.5µl *2
10x buffer
5µl
dNTP Mix
1µl
DMSO
1,25µl
H2O
36.25µl
sum
50µl
9-04-2010
9-05-2010
9-06-2010
name
4a-7
4a-8
4b-9
4b-10
3-3
3-6
6-11
6-12
DNA
4a; 2.4µl
4a; 2.4µl
4b; 0.5µl
4b; 0.5µl
3; 2µl
3; 2µl
6; 0.5µl
6; 0.5µl
primer [1pmol/µl]
7; 3.2µl
8; 3.2µl
9; 3.2µl
10; 3.2µl
3; 3.2µl
6; 3.2µl
11; 3.2µl
12; 3.2µl
Tris (10mM); pH 8.2
1.4µl
1.4µl
3.3µl
3.3µl
1.8µl
1.8µl
3.3µl
3.3µl
New overlapping PCR 5 with Pfu
PCR4a (~7ng)
0.56µl
PCR4b (~8ng)
1.1µl (1:10)
Pfu
0.5µl
Primer *2
2.5µl *2
10x buffer
5µl
dNTP Mix
1µl
DMSO
1,25µl
H2O
36.6µl
sum
50µl
New PCR 7a with Pfu
new dilution of 190-6; 1:100
template[2ng/µl]
1µl
2µl
pfu polymerase
0.5µl
0.5µl
primer13
2.5µl
2.5µl
primer14
2.5µl
2.5µl
buffer
5µl
5µl
dNTP Mix
1µl
1µl
DMSO
1,25µl
1.25µl
H2O
36.25µl
35.25
sum
50µl
50µl
Agarose gel electrophoresis of 7a
charges for sequencing for PCR 1,7b and 9
name
1-1
1-2
7b-15
7b-16
9-20
9-21
10-22
10-23
DNA
1[1:10]; 1.74µl
1[1:10]; 1.74µl
7b; 3µl
7b; 3µl
9; 1.78µl
9; 1.78µl
10; 3.8µl
10; 3.8µl
primer [1pmol/µl]
1; 3.2µl
2; 3.2µl
15; 3.2µl
16; 3.2µl
20; 3.2µl
21; 3.2µl
22; 3.2µl
23; 3.2µl
Tris (10mM); pH 8.2
2.06µl
2.06µl
0.8µl
0.8µl
2.02µl
2.02µl
0µl
0µl
9-07-2010
PCR nr.
1
2a
2b
3
4a
7b
9
10
expected band (bp)
492
332
772
1087
330
402
808
1888
shown band(s)
550,200
300
750
1100
300
450
900,1500
1900
clean charge
x
x
~x
x
x
x
Concentration
Low
High
template
1 µl (1:100)
15µl (1:100)
primer (11,12)
2.5 µl*2
dNTPs
1 µl
Pfu
0.5 µl
10xbuffer
5 µl
DMSO
1,25µl
H2O
36.25 µl
22.25 µl
sum
50µl
9-08-2010
eGFP
SV40PA=PCR6
ccdBamp
DNA
5µl=300ng
DNA
1µl=37,5ng
DNA
2µl=115ng
Buffer MC
2µl
Buffer D
2µl
Buffer H
2µl
BSA 1:10
2µl
BSA 1:10
2µl
BSA 1:10
2µl
EcoRI
0,5µl
XbaI
0,5µl
EcoRI
0,5µl
SpeI
0,5µl
PstI
0,5µl
PstI
0,5µl
H2O
13µl
H2O
14µl
H2O
13µl
sum
23µl
sum
20µl
sum
20µl
Ligation 1
eGFP
14,4µL
(144ng)
SV40PA
4,8µL
(48ng)
ccdBamp
2,9µL
(100ng)
T4 Buffer 10x
3µL
T4 Ligase
0,5µL
H2O
4,4µL
sum
30µL
Ligation at 22,5°C for 30 min, denaturation at 65°C for 10 min.
540ng
A260/A280=2,4
Transformation
9-09-2010
Plasmid extraction of ccdBcam, ccdBamp, Bak, CMV, PhiC31o
Plasmid
concentration [ng/µL]
A260/A280
ccdBcam
22.5
1.0
ccdBamp
42.5
1.2
Bak
17.5
0.8
CMV
10.0
0.7
PhiC310
60
1.4
PCRnr.
1
3
4a
4b
5
7a
9
10
template
pDS7 (1:100); 4µl
2a; 0.9µl+2b; 0.6µl
Bak; 0.5µl
Bak; 0.5µl
4a; 0.8µl+4b; 1µl
190-6 (1:100); 1µl
eGFP (1:25); 2µl
PhiC31o (1:10); 1µl
primer [10pmol/µl]
1,2; 2.5µl
3,6; 2.5µl
7,8; 2.5µl
9,10; 2.5µl
7,10; 2.5µl
13,14; 2.5µl
20,21; 2.5µl
22,23; 2.5µl
H2O
28µl
30.5µl
31.5µl
31.5µl
30.2µl
31µl
30µl
31µl
Annealing temperature
51.5°C
53.1°C
53.1°C
51.5°C
53.1°C
57.5°C
61°C
57.5°C
PCRnr.
1
4a
4b
5
7a
template
pDS7 (1:10); 1µl
Bak 1 ; 1µl
Bak 1; 1µl
4a, 4b; 2 x 1.5µl
pCT 190-6 (1:40); 1µl
primer
1, 2; 2 x 2.5µl
7, 8; 2 x 2.5µl
9, 10; 2 x 2.5µl
7, 10; 2 x 2.5µl
13, 14; 2x 2.5µl
H2O
31µl
31µl
31µl
30µl
30µl
Annealing temperature
52°C
52°C
48°C
50°C
55°C
9-10-2010
Agarose gel extraction of PCR 3, 4a, 5, 9, 10
PCR 3
PCR 4a
PCR 5
PCR 5*
PCR 9
PCR 10
concentration [ng/µl]
20
10
5
30
35
25
A260/A280
1.6
1.333
2.0
2.0
1.750
2.0
9-11-2010
9-12-2010
9-13-2010
name
4a-7
4a-8
4b-9
4b-10
3-3
3-6
6-11
6-12
DNA
4a; 2.4µl
4a; 2.4µl
4b; 0.5µl
4b; 0.5µl
3; 2µl
3; 2µl
6; 0.5µl
6; 0.5µl
primer [1pmol/µl]
7; 3.2µl
8; 3.2µl
9; 3.2µl
10; 3.2µl
3; 3.2µl
6; 3.2µl
11; 3.2µl
12; 3.2µl
Tris (10mM); pH 8.2
1.4µl
1.4µl
3.3µl
3.3µl
1.8µl
1.8µl
3.3µl
3.3µl
New PCR7a with Taq
template (~4ng)
2µl (1:100)
MasterMix for Taq
10µl
Primer *2
1.5µl *2
DMSO
0,5µl
H2O
4.5µl
sum
20µl
1: 94°C 2'
2: 94°C 30"
3: 64°C/62°C/60°C/58°C/56°C/54° 30"
4: 72°C 2'
5: (for each temperature)repeat 2-4 2x
6: 94°C 30"
7: 58°C 30"
8: 72°C 2'
9: repeat 6-8 29x
10: 72°C 10'
11: 15°C break
New PCR7a with Phusion Hot Start
template
2ng
4ng
H2O
54µl
53µl
Buffer 5x
20µl
20µl
Primer (13,14)
2*10µl
2*10µl
DMSO
3µl
3µl
Hot Start
1µl
1µl
9-14-2010
number:
7a diluted
7a undiluted
8
mastermix
50µl
50µl
10µl
primer
2*10µl
2*10µl
2*2µl
H2O
29µl
29µl
5µl
template
1µl 190-6 (1:100)
1µl 190-6
1µl 190-6 (1:100)
1
2
3
4
5
6
8
48°C
52°C
56,1°C
48°C
52°C
56,1°C
52°C
94°C 2'
94°C 30"
48°C/52°C/56,1°C 30"
72°C 1,5'
(for each temperature)repeat 30x
72°C 5'
9-15-2010
number
7a
7a
"8"
"8"
Mastermix
20µl
20µl
20µl
20µl
H2O
15µl
15µl
15µl
15µl
Primer
(13,14) 2*2µl
(13,14) 2*2µl
(13,16) 2*2µl
(13,16) 2*2µl
Template
190-6 (1:5) 1µl
190-6 (1:10) 1µl
190-6 (1:5) 1µl
190-6 (1:10) 1µl
program:
94°C 2'
94°C 30"
56°C/58°C 30"
72°C 1,5'
(for each temperature)repeat 30x
72°C 5'
Restriction Digestion of PCR 1 (17.8.), PCR 3 (10.9.), PCR 51 (10.9.), PCR 52 (10.9.), PCR 6 (17.8.), PCR 9 (10.9.), PCR 10 (30.8.) for ligation with vector
1
3
51
52
6
9
10
H2O
13µl
12,5µl
5µl
13µl
13,5µl
13,5µl
12,5µl
Buffer H
2µl
2µl
2µl
2µl
2µl
2µl
2µl
BSA (1:10)
2µl
2µl
2µl
2µl
2µl
2µl
2µl
DNA
2µl
2,5µl
10µl
2µl
1,5µl
1,5µl
2,5µl
EcoR1
0,5µl
0,5µl
0,5µl
0,5µl
0,5µl
0,5µl
0,5µl
Pst1
0,5µl
0,5µl
0,5µl
0,5µl
0,5µl
0,5µl
0,5µl
concentration (ng/µl)
A260/A280
(supposed) lenght
1
20
1,000
492
3
5
1,000
1087
51
22,5
1,8
688
52
7,5
1,5
688
6
10
1,333
237
9
20
1,6
808
10
17,5
1,75
1888
for
#
backbone
insert (µl)
charge (µl)
Buffer 10x (µl)
H2O (µl)
100ng
1
4
7,12
20
2
6,38
50ng
3
2
31,48
40
4
2,02
100ng
51
4
8,85
20
2
4,65
100ng
52
4
26,56
40
4
4,94
100ng
6
4
6,86
20
2
13,36
100ng
9
4
11,7
20
2
1,8
100ng
10
4
31,24
40
4
4,26
new PCR 7a & "8"
7a-1
7a-2
"8"-1
"8"-2
mastermix (µl)
10
10
10
10
template
190-6 1µl
190-6 2µl
190-6(1:10) 1µl
190-6(1:10) 2µl
primer
13&14 2*1µl
13&14 2*1µl
13&16 2*1µl
13&16 2*1µl
H2O
7µl
6µl
7µl
6µl
94°C 2'
94°C 30"
56°C 30"
72°C 1,5'
(for each temperature)repeat 30x
72°C 5'
12°C forever
9-16-2010
1
3
5
6
9
10
template
prev TRE(1:30) 1µl
2a(1:5) 1µl + 2b 0,5µl
4a 0,5µl + 4b 0,5µl
SV40PA (pcDNA3)(1:50) 1µl
eGFP(1:5) 1µl
PhiC310(1:6) 1µl
primer
1&2 2*2,5µl
3&6 2*2,5µl
7&10 2*2,5µl
11&12 2*2,5µl
20&21 2*2,5µl
22&23 2*2,5µl
Tm
54°C
50°C
49°C
56°C
58°C
58°C
mastermix (µl)
25
25
25
25
25
25
DMSO
1,25µl
1,25µl
1,25µl
1,25µl
1,25µl
1,25µl
H2O (µl)
17,75
17,25
17,75
17,75
17,75
17,75
94°C 2'
94°C 30"
Tm (see above) 30"
72°C 1,5'
(for each temperature)repeat 30x
72°C 5'
9-17-2010
1
3
5
6
9
10
pellet
no
yes
yes
yes
yes
yes
yes
100µl
no
yes
yes
yes
yes
yes
yes
1
5
6
template
pTRE Rev 0,5µl
4a 1,5µl + 4b 1,5µl
pcDNA3 0,5µl
primer
1&2 2*2,5µl
7&10 2*2,5µl
11&12 2*2,5µl
buffer 10x
5µl
5µl
5µl
dNTPs
1µl
1µl
1µl
Pfu
0,5µl
0,5µl
0,5µl
H2O
26,75 µl
24,25 µl
26,75 µl
T<sub<m</sub>
51°C
50°C
55°C
Restriction digestion of PCR products, ccdB Plasmids and Biobricks for the 3A Method
Name
H2O
Buffer (each 2µl)
BSA (1:10)
DNA Volume
DNA Mass
Enzyms (2*0.5µl)
ccdBa
25 µl
H
2µl
20µl
~600ng
E+P
ccdBa
25µl
H
2µl
20µl
~700ng
E+P
1
43µl
MC
2µl
2µl
~500ng
E+S
3
25µl
B
2µl
20µl
~200ng
X+P
Primer 18+19
25µl
B
2µl
10µl+10µl
?
X+P
51
25µl
MC
2µl
20µl
~100ng
E+S
52
25µl
MC
2µl
20µl
~600ng
E+S
6
35µl
B
2µl
10µl
~400ng
X+P
9
25µl
MC
2µl
20µl
~700ng
E+S
10
25µl
MC
2µl
20µl
~400ng
E+S
eGFP
35µl
MC
2µl
10µl
~550ng
E+S
CMV1
35µl
H
2µl
10µl
~530ng
E+P
PCR Mastermix
10µl
Biobrick Primer F
1.5µl
Biobrick Primer R
1.5µl
H2O
7µl
PCR Mastermix
10µl
Biobrick Primer F
1.5µl
Biobrick Primer R
1.5µl
Template
2µl
H2O
5µl
PCR clean up of PCR Gel extraction 31, 32, 5, 9, 10, PCR Product 6 and digestion PCR1, Primer 18+19, R51, R52, PCR6, PCR9, PCR10, PCR3
9-18-2010
9-19-2010
9-20-2010
9-21-2010
PCR 7a with new Primers
template 190-6 (1:10)
1µl
Master Mix
25µl
Primer 13k (short)
3,75µl
Primer 14
3,75µl
H2O
16,5µl
key: JD=Jump-or-Die; CS=Cut'N'Survive; first number=Ligation number (see schedule); second number=colonie number (marked on the plate)
verified products:7a (~800bp); JD.2.1; JD.2.2(each ~1000bp);JD.31.1(~2000bp); CS.1a.2(~1500bp)
9-22-2010
PCR Mastermix
55µl
Primer F
8,25µl
Primer R
8,25µl
H2O
38,5µl
sum
110µl
PCR Purification of 7a
PCR8 with Phusion Hot Start II
template 7a 1:100
3µl ~4ng (850bp)
template 7b 1:10
2µl ~2ng (402bp)
Buffer 5X
10µl
Phusion
0.5µl
H2O
27µl
Primer 13
2.5µl
Primer 16
2.55µl
DMSO)
1.5µl
dNTP mix
1µl
Plasmid extraction of "over-day" cultures ligation-7(=BB4),-8(=BB4),-9(=BB5),-12(=BB9) and pSB1C3
Master Mix
25µl
H2O
16,5µl
Primer 15
3,75µl
Primer 16
3,75µl
template 190-6 (1:10)
1µl
9-23-2010
Name
H2O
Buffer D
BSA (1:10)
DNA Volume
DNA Mass
Enzyms (2*0.5µl)
BB5
11 µl
2µl
2µl
4µl
~500ng
X+P
Name
H2O
Buffer D
BSA (1:10)
DNA Volume
DNA Mass
Enzyms (2*0.5µl)
BB2
10 µl
2µl
2µl
5µl
~1.2mg
X+P
Name
H2O
Buffer D
BSA (1:10)
DNA Volume
DNA Mass
Enzyms (2*0.5µl)
BB11
7 µl
2µl
2µl
8µl
~1.3mg
X+P
Ligation of BB1 and BB6
Name
H2O
T4 Ligase-Buffer
PCR1
Primer 18+19
ccdBAmp
T4 Ligase
BB1
9.5 µl
2µl
5µl
2µl
1µl
0.5µl
Name
H2O
T4 Ligase-Buffer
CMV E+S
BB5
ccdBCam
T4 Ligase
BB6
6.5 µl
2µl
8µl
2µl
1µl
0.5µl
Buffer 5x
10µl
H2O
27µl
dNTPs
1µl
Primer 16
2,5µl
Primer 13k short
2,5µl
DMSO
1,5µl
template 7a(1:100)(850bp)
3µl -> 4ng
template 7b(new)(1:100)(402bp)
2µl -> 2ng
Phusion Hot Start II
0,5µl
BB1 plated on amp-Agar, BB6 and pSB1C3 plated on cam-Agar
9-24-2010
PCR Mastermix 2x
10µl
Primer F
1.5µl
Primer R
1.5µl
H2O
7µl
sum
20µl
concentration
82.5ng/µl
A260/A280
1.737
Agarose Gelelektrophoresis of 11, 12, 61 and 62
Plasmid extraction of pSB1C3 in cam vector
concentration
22.5ng/µl
A260/A280
1.5
9-25-2010
9-26-2010
9-27-2010
PCR Mastermix
17.5µl
Primer forward
2.625µl
Primer revers
2.7µl
H2O
12.1µl
sum
35µl = 5µl per charge
Ligation of BB10
CMV© E+S
5µl
PCR 8l X+P
5µl
ccdBamp
1µl
T4 Ligase-Buffer
2µl
T4 Ligase
0.5µl
H2O
6.5µl
sum
20µl
Ligation of BB001(PCR1 in ©), BB017(PCR5 in ©), BB018(PCR6 in ©) and BB021(PCR9 in ©)
ccdB
1µl
BB001/017/018/021
5µl
T4 Ligase-Buffer
2µl
T4 Ligase
0.5µl
H2O
11.5µl
sum
20µl
template
H2O
BSA [1:10]
Buffer H
EcoRI
PstI
PCR32
10µl
5µl
2µl
2µl
0.5µl
0.5µl
PCR8l
10µl
5µl
2µl
2µl
0.5µl
0.5µl
Primer 18+19
2x7.5µl
--
2µl
2µl
0.5µl
0.5µl
PCR10
10µl
5µl
2µl
2µl
0.5µl
0.5µl
9-28-2010
Amp
BB72
BB10
Cam
BB001
BB017
BB018
BB021
template
pick up colony from plate
primers*2
0.25µl*2
dNTPs
0.5µl
DreamTaq
0.05µl
10xbuffer
0.5µl
H2O
3.45µl
sum
5µl
BB2
BB019
BB020
BB016
BB022
Volume
PCR
52
8
32
10
each 5µl
PCR
6
each 5µl
Primer
18+19
each 5µl
ccdBamp
1µl
ccdBcam
1µl
1µl
1µl
1µl
T4 Ligase-Buffer
2µl
2µl
2µl
2µl
2µl
T4 Ligase
0.5µl
0.5µl
0.5µl
0.5µl
0.5µl
H2O
6.5µl
11.4µl
11.4µl
11.4µl
11.4µl
sum
20µl
20µl
20µl
20µl
20µl
9-29-2010
H2O
6,7µl
Buffer 10x
1µl
MgCl2
1µl
dNTPs
0,2µl
Primer forw. seq f
0,5µl
Primer rev. seq r
0,5µl
Taq
0,1µl
1
2
3
4
5
6
7
8
9
10
BB2/BB8 - 1
BB2/BB8 - 2
BB016 - 1
BB016 - 2
BB019 - 1
BB019 - 2
BB020 - 1
BB020 - 2
BB022 - 1
BB022 - 2
PCR (5 µl each)
PCR (5µl each)
ccdB(amp)
ccdB©
Buffer
T4 Ligase
H2O
BB72O
1
32
1µl
2µl
0,5µl
6,5µl
BB017
51
1µl
2µl
0,5µl
11,5µl
BB10
CMV©
PCR8
1µl
2µl
0,5µl
6,5µl
BB021
9
1µl
2µl
0,5µl
11,5µl
BB001
1
1µl
2µl
0,5µl
11,5µl
BB018
6
1µl
2µl
0,5µl
11,5µl
template
digestion with
volume
PCR 6
Xba1 & Pst1
I. 10µl II. 3µl
PCR52
EcoR1 & Pst1
20µl
PCR32
Xba1 & Pst1
10µl
PCR1
EcoR1 & Spe1
2µl
ccdB(amp)
EcoR1 & Pst1
20µl
cddB(cam)
EcoR1 & Pst1
20µl
colony-PCR of BB4/c3, BB6/c7, BB72/c5, BB1/c5, (BB11 as control) with Taq
H2O
32µl
Buffer 10x
5µl
MgCl2
5µl
dNTPs
1µl
Primer forw. seq f
2,5µl
Primer rev. seq r
2,5µl
Taq
0,5µl
DMSO
1,5µl
sum
50µl
1
2
3
4
5
BB4-c3
BB6-c7
BB72-c5
BB1-c5
BB11
9-30-2010
Colony/BB11
2.5µl
H2O
32µl
dNTP
1µl
Primer F
2.5µl
Primer R
2.5µl
MgCl2
5µl
DMSO
1.5µl
Buffer 10x (with KCl)
5µl
Taq Polymerase
0.5µl
sum
56µl
concentration [ng/µl]
A260/A280
6II
32.5
1.444
ccdBamp
0.0
---
ccdBcam
5.0
1.000
6I
0.0
---
52
20.0
1.143
32
27.5
1.57
1
25
1.667
concentration [ng/µl]
A260/A280
BB020(K1)
163
1.711
BB018(K5)
92.5
1.85
BB6(K5)
120
1.778
BB022(K1)
105
1.75
BB016(K1)
85
1.889
BB019(K1)
105
1.909
BB2(K1)
190
1.81
BB018
110
1.833
10-01-2010
H2O
67µl
dNTP
2µl
Primer F
5µl
Primer R
5µl
MgCl2
10µl
Buffer 10x (with KCl)
10µl
Taq Polymerase
1µl
sum
100µl
10-02-2010
10-03-2010
10-04-2010
CMV (c)
PCR 1 (c)
BB4 7
BB4 8
BB018K5
BB018K3
BB6K5
BB2K1
template
1µl
2µl
3µl
3,8µl
2µl
2µl
2µl
1µl
primer (1:100)
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
10mM TrispH 8,5
2,8 µl
1,8 µl
0,8 µl
0 µl
1,8 µl
1,8 µl
1,8 µl
1,8 µl
name of tube
028 FIR
001 F/R
4-7 F/R
4-8 F/R
018-5 F/R
018-3 F/R
6-5 F/R
2-1 F/R
PCR
019
022
016
020
template (1:100 = 2ng)
2µl
2µl
2,5µl
1,5µl
Primer F
1µl
1µl
1µl
1µl
Primer R
1µl
1µl
1µl
1µl
dNTPs
0,4µl
0,4µl
0,4µl
0,4µl
Taq
0,2µl
0,2µl
0,2µl
0,2µl
Buffer
2µl
2µl
2µl
2µl
MgCl2
2µl
2µl
2µl
2µl
H2O
11,4µl
11,4µl
10,9µl
11,
colour of tube
orange
rose
green
blue
10-05-2010
gel 1
BB6K9
BB6K10
BB6K11
BBK12
BB6K13
BB4K5
ladder
BB4K6
BB4K7
BB021K14
BB021K13
BB021K12
BB021K11
BB022K18
BB022K19
ladder
BB022K17
BB022K16
gel 2
BB022K12
BB022K11
CMV©K4
CMV©K5
BB001K5
BB001K6
BB001K7
BB001K8
ladder
BB016K17
BB1K16
BB1K15
BB1K8
BB1K7
BB252K1
BB252K2
BB252K3
ladder
gel 3
BB017K14
BB017K13
BB017K12
BB017K11
BB018K17
BB018K16
BB018K15
BB018K14
ladder
BB019K6
BB019K5
BB019K4
ladder
BB019K3
BB020K7
BB020K8
BB020K6
BB020K5
gel 1
BB4K8
BB4K9
BB4K10
BB021K10
BB021K15
BB022K15
BB022K14
BB022K13
---
---
gel 2
BB001K9
BB001K10
BB016K11
BB016K12
BB016K13
BB016K14
BB016K15
BB016K16
---
---
BB252K4
BB252K5
BB72K12
BB72K11
BB72K10
BB72K9
BB017K16
BB017K15
gel 3
BB018K13
BB018K12
BB018K11
BB018K10
BB018K9
BB020K4
BB020K3
---
---
10-06-2010
concentration [ng/µl]
A260/A280
BB6(K13)
265
1.8
BB4(K10)
40
2.0
CMVc(K4)
97.5
1.8
BB001(K5)
30
1.7
BB001(K9)
17.5
1.7
BB016(K11)
45
1.7
BB1(K8)
57.5
1.9
BB2(K3)
60
1.8
BB7(K10)
30
1.7
BB017(K14)
12.5
1.5
BB018(K10)
35
1.8
BB019(K6)
37.5
1.5
10-07-2010
Primer fw
0.5 µl
Primer rev
0.5 µl
dNTPs
0.2 µl
Taq
0.1 µl
10xbuffer
1 µl
MgCl2
1 µl
H2O
6.7 µl
10-08-2010
concentration [ng/µl]
A260/A280
BB4(K10)
258
1,981
BB001(K5)
325
1.94
BB001(K9)
205
1.907
BB7(K10)
170
1.744
BB017(K14)
215
1.955
BB018(K10)
500
1.88
BB019(K6)
293
1.95
10-09-2010
10-10-2010
10-11-2010
concentration [ng/µl]
A260/A280
BB018K5
55
1.571
BB018K6
92,5
1.762
BB021K24
27.5
1.833
BB021K17
37.5
1.508
Colony PCR BB01, BB018, BB6, BB252
stripe 1
BB001K5
K6
K7
K8
K9
K9
K10
BB252 K2
stripe 2
BB252 K3
K4
K5
BB6 K9
K10
K11
K12
K13
stripe 3
BB018 K9
K10
K11
K12
K13
K14
K15
K16
single sample
BB018 K17
gel picture colony PCR
BB001K9 for
BB001K9 rev
BB4K16 for
BB4K16 rev
BB001K5 for
BB001K5 rev
BB252K3
BB252K3
template
1µl
1µl
1µl
1µl
0,5µl
0,5µl
3,8µl
3,8µl
primer (1:100)
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
10mM TrispH 8,5
2,8 µl
2,8 µl
2,8 µl
2,8 µl
3,3 µl
3,3 µl
0 µl
0 µl
name of tube
31-F
31-R
44-F
44-R
24-F
24-R
32-F
32-R
BB72K10 for
BB72K10 rev
BB017K14 for
BB017K14 rev
BB018K10 for
BB018K10 rev
BB019K6 for
BB019K6 rev
template
1,5µl
1,5µl
1µl
1µl
0,5µl
0,5µl
0,8µl
0,8µl
primer (1:100)
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
10mM TrispH 8,5
2,3 µl
2,3 µl
2,8 µl
2,8 µl
3,3 µl
3,3 µl
3 µl
3 µl
name of tube
14-F
14-R
21-F
21-R
13-F
13-R
34-F
34-R
CMV for
CMV rev
BB018K5 for
BB018K5 rev
BB018K6 for
BB018K6 rev
BB021K17
BB021K17
template
2µl
2µl
3,8µl
3,8µl
2µl
2µl
3,8µl
3,8µl
primer (1:100)
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
3,2 µl
10mM TrispH 8,5
1,8 µl
1,8 µl
0 µl
0 µl
1,8 µl
1,8 µl
0 µl
0 µl
name of tube
CMV-F
CMV-R
185-F
185-R
186-F
186-R
2117-F
2117-R
ligation BB020 BB022, Bb6 and BB52
BB020
BB022
BB6
BB52
insert (5µl)
18+19
PCR 10
CMV
PCR5<sub<2</sub>
insert (5µl)
-
-
BB5
PCR6
ccdB (1µl)
c
c
c
a
buffer
2µl
2µl
2µl
2µl
T4-ligase
0,5µl
0,5µl
0,5µl
0,5µl
H2O
11,5µl
11,5µl
6,5µl
6,5µl
template eGFP
1µl
Primer 20,21
each 1µl
buffer (10x)
2µl
MgCl
2µl
dNTPs
0,2µl
taq
0,3µl to 0,7µl (0,1µl steps)
H2O
fill to 20 µl
![]()
![]()

 "
"