Team:Freiburg Bioware/NoteBook/Labjournal/October2
From 2010.igem.org
(→Concentration of virus stock for Western Blot analysis) |
|||
| (274 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Freiburg_Bioware/Head}} | {{:Team:Freiburg_Bioware/Head}} | ||
{{:Team:Freiburg_Bioware/css}} | {{:Team:Freiburg_Bioware/css}} | ||
| + | {{:Team:Freiburg_Bioware/menu_notebook}} | ||
| + | {{:Team:Freiburg_Bioware/jquery}} | ||
<!-- Freiburg_bioware --> | <!-- Freiburg_bioware --> | ||
| + | [https://2010.igem.org/Team:Freiburg_Bioware/NoteBook => Back to Notebook overview]<br><br> | ||
<html> | <html> | ||
| Line 22: | Line 25: | ||
<li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/September">September part 1 (labday 107 - 123)</a></li> | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/September">September part 1 (labday 107 - 123)</a></li> | ||
<li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/September2">September part 2 (labday 124 - 135)</a></li> | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/September2">September part 2 (labday 124 - 135)</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/October">October part 1 (labday 136 - | + | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/October">October part 1 (labday 136 - 149 )</a></li> |
| - | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/October2">October part 2 (labday | + | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/October2">October part 2 (labday 150 - 166 )</a></li> |
| - | + | ||
<li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/November">November (labday 167 - 170 )</a></li> | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/November">November (labday 167 - 170 )</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/Cellculture">Cellculture </a></li> | + | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/Cellculture">Cellculture</a></li> |
</ul> | </ul> | ||
</div> | </div> | ||
| Line 33: | Line 35: | ||
<!-- NoteBook --> | <!-- NoteBook --> | ||
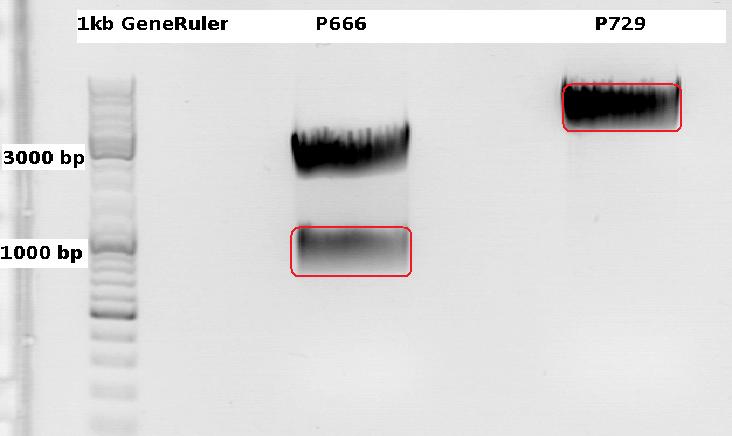
| - | ===<p style="font-size:17px; background-color:#00dd77;"> | + | ===<p style="font-size:17px; background-color:#00dd77;">150. labday 15.10.2010</p>=== |
| - | ====<p style="font-size:15px; background-color:#66bbff;"> | + | ====<p style="font-size:15px; background-color:#66bbff;">Cloning CFP (from P666: PSB1C3_CFP) into pSB1C3_leftITR_CMV_beta-globin (P729)</p>==== |
| - | + | '''Investigator Patrick <br> | |
| - | + | Digestions, 2 h 10 minutes, 37 °C: | |
| - | + | *P666: 5 µl DNA, 2 µl BSA, 2 µl Buffer 4 (10x), 1 µl Xba, 1 µl PstI, 9 µl H2O | |
| - | + | *P729: 4 µl DNA, 2 µl BSA, 2 µl Buffer 4 (10x), 1 µl SpeI, 1 µl PstI, 10 µl H2O | |
| - | + | ||
| - | + | ||
| - | + | Expected results for the 1% agarose gel: | |
| + | *P666: about 2100 and 750 bp | ||
| + | *P729: about 3300 and 20 bp | ||
| - | [[Image: | + | <br> |
| + | [[Image:Freiburg10_1015pat_fertig.jpg|thumb|center|600px]] | ||
| + | <br> | ||
| + | <br> | ||
| + | The gelextraction ... | ||
| + | |||
| + | P728: 11,8 ng/µl | ||
| + | <br> | ||
| + | P822: 34,6 ng/µl | ||
| + | |||
| + | ... and following ligation (2,5 µl Insert, 5,5 µl vector, 1 µl T4 DNA Ligase, 1 µl T4 DNA Ligase Buffer (10x), 40 minutes, RT) were performed according to the standard protocol. After the transformation (with XL1B) the cells were plated and put into the 37°C room. | ||
| + | Two additional transformations were performed with ligations from Volker labeled: "ligation viral brick 453 empty" & "viral brick 587 empty". <br> | ||
| + | |||
| + | The following day the plates were checked for clones. Unfortunately there grew no clones on these two plates contrary to my plate with a a lot of clones. | ||
| + | |||
| + | ====<p style="font-size:15px; background-color:#66bbff;">Midi-Prep</p>==== | ||
| + | |||
| + | '''Investigator: Chris W. <br>''' | ||
| + | <p style="font-size:13px; color:#003399;"> Midi-Prep of:</p><br/> | ||
| + | pSB1C3_001_RC_IRCK_P5tataless clone 1 =P866 =B516<br/> | ||
| + | pSB1C3_001_CMV_VP123_587-KO_Z34C_spacer clone2 =P867 =B526<br/> | ||
| + | pSB1C3_001_CMV_VP123_587-KO_Z34C clone2 =P868 =B529<br/> | ||
| + | pSB1C3_CMV_Zegfr:1907_MiddleLinker_VP2/3_587-KO_BAP clone 1 =P869 =B680<br/> | ||
| + | pSB1C3_CMV_Zegfr:1907_MiddleLinker_VP2/3_587-KO_6xHis clone 1 =P870 =B200<br/> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br/> | <br/> | ||
| + | The Midi-Preps were performed according to the standard protocol yielding the following concentrations: | ||
| - | + | {| border="1" | |
| + | | plasmid-no. || align="right" |P866|| align="right" |P867|| align="right" |P868|| align="right" |P869|| align="right" |P870 | ||
| + | |- | ||
| + | | concentration (ng/µl)|| align="right" |899,80 || align="right" |954,46 || align="right" |406,97|| align="right" |1642,76|| align="right" |1585,12 | ||
| + | |} | ||
| + | <br> | ||
<br/> | <br/> | ||
| - | + | ====<p style="font-size:15px; background-color:#66bbff;">mini prep of several constructs</p>==== | |
| - | + | '''Investigator: Kira <br/> | |
| - | + | ||
| - | + | c(rep52_1)=299,04 ng/ul<br/> | |
| - | + | c(rep52_2)=290,07 ng/ul<br/> | |
| - | + | c(rep78_1)=142,32 ng/ul<br/> | |
| + | c(rep78_2)=175,36 ng/ul<br/> | ||
| + | |||
| + | ====<p style="font-size:15px; background-color:#66bbff;">Cell culture</p>==== | ||
| + | '''Investigator: Kira <br/> | ||
| + | |||
| + | The cells are still alive. Medium was exchanged.--> RNA will be harvested tomorrow | ||
| + | |||
| + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Mini-Prep and test digestion of pSB1C3_CD_SDM-PstI_hGH_rITR</b></p>==== | ||
| + | <b>Investigator: Stefan</b><br /> | ||
| + | |||
| + | Glycerol stocks were prepared:<br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
<ul> | <ul> | ||
| - | <li> | + | <li>B694 = pSB1C3_CD_SDM-PstI_hGH_rITR clone 1</li> |
| - | <li> | + | <li>B695 = pSB1C3_CD_SDM-PstI_hGH_rITR clone 2</li> |
| + | |||
| + | </ul><br> | ||
| + | |||
| + | Mini-Prep was performed according to standard protocol:<br> | ||
| + | |||
| + | <ul> | ||
| + | <li>P875 = pSB1C3_CD_SDM-PstI_hGH_rITR clone 1 c = 73,1 ng/µl</li> | ||
| + | <li>P876 = pSB1C3_CD_SDM-PstI_hGH_rITR clone 2 c = 78,3 ng/µl</li> | ||
</ul> | </ul> | ||
| - | [[Image: | + | |
| + | Test digestion:<br> | ||
| + | {| border="1" | ||
| + | | align="left" | '''Components''' ||align="left"| <b>P875 + P876 / µl</b> | ||
| + | |- | ||
| + | | align="left" | DNA ||align="left"| 4 | ||
| + | |- | ||
| + | | align="left" | Buffer 4 ||align="left"| 1 | ||
| + | |- | ||
| + | | align="left" | BSA (10x) ||align="left"| 1 | ||
| + | |- | ||
| + | | align="left" | XbaI ||align="left"| 0,3 | ||
| + | |- | ||
| + | | align="left" | AgeI ||align="left"| 0,3 | ||
| + | |- | ||
| + | | align="left" | H<sub>2</sub>O ||align="left"| 3,4 | ||
| + | |- | ||
| + | | align="left" | '''Total volume''' ||align="left"| <b>10</b> | ||
| + | |} | ||
| + | |||
| + | Gel:<br> | ||
| + | 0,5g agarose, 50 ml TAE (1%), 3 µl GELRED, 115 Volt, running time ~50 minutes<br> | ||
| + | |||
| + | [[Image:Freiburg10 td151010.jpg|550px|thumb|center]] | ||
| + | <p style="color:#66bbff;"><i>Comment</i>: Test digestion looks allright, cloning will be continued using P876.</p> | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">151. labday 16.10.2010</p>=== | ||
| + | ====<p style="font-size:15px; background-color:#66bbff;">Biobrick assembly: pSB1C3_lITR_CMV_ß-globin_CD_hGH_rITR and pSB1C3_lITR_phTERT_ß-globin_CD_hGH_rITR </p>==== | ||
| + | |||
| + | '''Investigator: Achim''' | ||
| + | |||
| + | '''Plasmids:''' | ||
| + | *P729: pSB1C3_lITR_CMV_ß-Globin | ||
| + | **c= 243.4 ng/µl | ||
| + | *P730: pSB1C3_lITR_pHTERT_ß-Globin | ||
| + | **c= 81.1 ng/µl | ||
| + | *P876: pSB1C3_CD_SDM-PstI_hGH_rITR | ||
| + | **c= 78.3 ng/µl | ||
| + | '''Digestion:''' | ||
| + | |||
| + | {| border="1" | ||
| + | | <b>components</b> || align="right" |<b>I1 (P729)</b> || align="right" |<b>I2 (P730)</b> || align="right" |<b>V (P876) | ||
| + | |- | ||
| + | | DNA || align="right" |6 || align="right" |14|| align="right" |14 | ||
| + | |- | ||
| + | | BSA (10x) || align="right" |2 || align="right" |2|| align="right" |2 | ||
| + | |- | ||
| + | | Buffer 4 (10x)|| align="right" |2 || align="right" |2 || align="right" |2 | ||
| + | |- | ||
| + | |Enzyme EcoI|| align="right" |1|| align="right" |1 || align="right" |1 | ||
| + | |- | ||
| + | |Enzyme XbaI|| align="right" |-|| align="right" |- || align="right" |1 | ||
| + | |- | ||
| + | |Enzyme SpeI|| align="right" |1|| align="right" |1 || align="right" |- | ||
| + | |- | ||
| + | |H2O|| align="right" |8|| align="right" |- || align="right" |- | ||
| + | |- | ||
| + | |'''Total '''|| align="right" | 20|| align="right" | 20|| align="right" | 20 | ||
| + | |} | ||
| + | |||
| + | Digestion: 2h, 37°C | ||
| + | |||
| + | '''Prep. gel:''' | ||
| + | *0,8%, run for 45 min | ||
| + | |||
| + | [[Image:Freiburg10 1610 bba.png|thumb|none|400px|Expected Bands: I1: 1335, I2: 1138, V: 4000]] | ||
| + | |||
| + | *Corresponding bands were cut out | ||
| + | |||
| + | '''Gel ex.''' | ||
| + | |||
| + | *Nanodrop concentrations: | ||
| + | **I1: 37.54 ng/µl | ||
| + | **I2: 25.38 ng/µl | ||
| + | **V: 26.47 ng/µl | ||
| + | |||
| + | '''Ligation:''' | ||
| + | {| border="1" | ||
| + | |ligation name || align="right" |I1 + V|| align="right" |I2 + V | ||
| + | |- | ||
| + | |volume of vector || align="right" |4.69|| align="right" |4.23 | ||
| + | |- | ||
| + | |volume of insert|| align="right" |3.31|| align="right" |3.77 | ||
| + | |- | ||
| + | |T4 ligase buffer (10x)|| align="right" |1|| align="right" |1 | ||
| + | |- | ||
| + | |T4 ligase || align="right" |1|| align="right" |1 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | *Ligation @ RT for 40 min | ||
| + | |||
| + | '''Trafo:''' | ||
| + | *Done by Kira | ||
| + | |||
| + | |||
| + | <br> | ||
| + | |||
| + | ====<p style="font-size:15px; background-color:#66bbff;">Mini-prep of mutual pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_hGH_rITR_SDM-PstI</p>==== | ||
| + | |||
| + | '''Investigator Patrick | ||
| + | <br> | ||
| + | |||
| + | Yielded concentrations & given numbers: | ||
| + | *pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_hGH_rITR_SDM-PstI clone 1: 208,4 ng/µl , P877 / B696 | ||
| + | *pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_hGH_rITR_SDM-PstI clone 2: 251,6 ng/µl , P878 / B696 | ||
| + | *pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_hGH_rITR_SDM-PstI clone 3: 212,6 ng/µl , P889 / B696 | ||
| + | |||
| + | |||
| + | |||
| + | <br> | ||
| + | Test-digestion: 0,5 µl SpeI, 0,5 µl PstI, 3 µl DNA, 1 µl Buffer 4, 1 µl BSA, 4 µl H2O, 40 minutes, 37°C | ||
| + | <br> | ||
| + | Expected results: fragments with about 650 and 4900 bp | ||
| + | <br> | ||
| + | [[Image:Freiburg10_1016pat_test.jpg|thumb|center|600px]] | ||
| + | <br> | ||
| + | Obviously, this test digestion has to be repeated. | ||
| + | |||
| + | Preparations for tomorrow: | ||
| + | *Mini-prep of 3 mutual pSB1C3_leftITR_CMV_beta-globin_CFP clones (have to be picked from the plate) | ||
| + | *Midi-prep of pHelper | ||
| + | *Midi-prep of B689:pSB1C3_lITR_CMV_betaglobin_mGMK_TK30_SDM-PstI_hGH_rITR clone 2 | ||
| + | |||
| + | ====<p style="font-size:15px; background-color:#66bbff;">Biobrick assembly of Rep78 and Rep52</p>==== | ||
| + | |||
| + | '''Investigator: Kira <br /> | ||
| + | '''Comment:''' After replacing the mutated Rep parts by the ordered Rep parts, PCR amplification has to be done in order to produce a biobrick. | ||
| + | PCR program: | ||
<br /> | <br /> | ||
| - | + | ||
| + | c(Rep52)=299 ng/ul <br /> | ||
| + | c(Rep78)=175 ng/ul <br /> | ||
| + | |||
| + | Rep52: praefix 094 & suffix 097 <br /> | ||
| + | Rep78: praefix 093 & suffix 097 <br /> | ||
| + | |||
| + | |||
| + | {| border="1" | ||
| + | | components || align="right" |volume in µl | ||
| + | |- | ||
| + | | 5x Phusion HF buffer || align="right" | 10 | ||
| + | |- | ||
| + | | 10 mM dNTP mix || align="right" |1 | ||
| + | |- | ||
| + | |primer_for (1:10 dilution)|| align="right" | 2,5 | ||
| + | |- | ||
| + | |primer_rev (1:10 dilution)|| align="right" | 2,5 | ||
| + | |- | ||
| + | |DNA template (1:100)|| align="right" | 0,5 | ||
| + | |- | ||
| + | |DMSO|| align="right" | 0,5 | ||
| + | |- | ||
| + | |Phusion polymerase|| align="right" | 0,5 | ||
| + | |- | ||
| + | |H<sub>2</sub>O|| align="right" | 32,5 | ||
| + | |- | ||
| + | |'''Total volume (e.g. 50 µl)'''|| align="right" | 50 | ||
| + | |} | ||
<br /> | <br /> | ||
| - | < | + | |
| + | {| border="1" | ||
| + | |Cycles||Temperature||Time | ||
| + | |- | ||
| + | |||98°C||30 sec | ||
| + | |- | ||
| + | |10x||98°C||15 sec | ||
| + | |- | ||
| + | |||63°C||25 sec | ||
| + | |- | ||
| + | |||72°C||32 sec | ||
| + | |- | ||
| + | |20x||98°C||15 sec | ||
| + | |- | ||
| + | |||66°C||25 sec | ||
| + | |- | ||
| + | |||72°C||32 sec | ||
| + | |- | ||
| + | |1x||72°C||5 min | ||
| + | |- | ||
| + | |Hold 4°C | ||
| + | |} | ||
| + | <br> | ||
| + | 1% agarose gel <br /> | ||
| + | [[Image:Freiburg10_Rep78&Rep52_PCR.jpg]] | ||
| + | |||
| + | Digestion of plasmid backbone: | ||
| + | |||
| + | pSB1C3_001 is used as backbone | ||
| + | |||
| + | {| border="1" | ||
| + | | align="left" | '''Components''' ||align="left"| <b>vector</b> Volume/µL | ||
| + | |- | ||
| + | | align="left" | DNA ||align="left"| 3,5 µl | ||
| + | |- | ||
| + | | align="left" | BSA (10x) ||align="left"| 2 µl | ||
| + | |- | ||
| + | | align="left" | Buffer no. 4 (10x) ||align="left"| 2,0 µl | ||
| + | |- | ||
| + | | align="left" | Enzyme 1 EcoRI-HF ||align="left"| 0,5 µl | ||
| + | |- | ||
| + | | align="left" | Enzyme 2 SpeI ||align="left"| 1,0 µl | ||
| + | |- | ||
| + | | align="left" | H<sub>2</sub>O ||align="left"| 15 µl | ||
| + | |- | ||
| + | | align="left" | '''Total volume''' ||align="left"| <b>25</b> | ||
| + | |} | ||
<br /> | <br /> | ||
| - | [[Image: | + | |
| + | incubation @ 37 C for approx. 2 h | ||
| + | |||
| + | 1% agarose gel <br /> | ||
| + | [[Image:Freiburg10_digestion_pSB1C31_16.10.jpg|thumb|center|600px]] | ||
| + | |||
| + | Digestion of PCR product: | ||
| + | {| border="1" | ||
| + | | align="left" | '''Components''' ||align="left"| <b>PCR product</b> Volume/µL | ||
| + | |- | ||
| + | | align="left" | DNA ||align="left"| 35,0 µl | ||
| + | |- | ||
| + | | align="left" | BSA (100x) ||align="left"| 0,45 µl | ||
| + | |- | ||
| + | | align="left" | Buffer no. 4 ||align="left"| 4,5 µl | ||
| + | |- | ||
| + | | align="left" | Enzyme 1 EcoRI-HF ||align="left"| 1,5 µl | ||
| + | |- | ||
| + | | align="left" | Enzyme 2 SpeI ||align="left"|2,0 µl | ||
| + | |- | ||
| + | | align="left" | H<sub>2</sub>O ||align="left"| 1,5 µl | ||
| + | |- | ||
| + | | align="left" | '''Total volume''' ||align="left"| <b>45</b> | ||
| + | |} | ||
<br /> | <br /> | ||
| + | incubation @ 37 C for approx. 2 h <br /> | ||
| + | |||
| + | T4 ligation for 40 min <br /> | ||
| + | Transformation according to the standard protocol <br /> | ||
| + | |||
| + | ====<p style="font-size:15px; background-color:#66bbff;">RNA harvesting</p>==== | ||
| + | |||
| + | '''Investigator: Kira <br /> | ||
| + | After transfection, the cells were incubated for 48 hours. Today, the cells will be harvested and RNA extracted, in order to perform RT-PCR and an additional PCR for evaluation of promoter activity.<br /> | ||
| + | |||
| + | The transfected cells were trypsinised and centrifuged for 2 min. The supernatant was discarded and pellet washed 2x with PBS. RNeasy Kit [Qiagen] was used for RNA extraction according to the manufacturer protocol. <br /> | ||
| + | |||
| + | c(CMV)= 335,69 ng/ul<br /> | ||
| + | c(P40)= 857,92 ng/ul<br /> | ||
| + | c(AAV_RC)= 760,21 ng/ul<br /> | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">152. labday 17.10.2010</p>=== | ||
| + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Test digestion of pSB1C3_lITR_CMV_ß-globin_CFP</b></p>==== | ||
| + | <b>Investigator: Anna</b><br> | ||
| + | |||
| + | Vector name:<br /> | ||
| + | pSB1C3_lITR_CMV_betaglobin_CFP_cl1 (P880): c = 452,67 ng/µl<br /> | ||
| + | pSB1C3_lITR_CMV_betaglobin_CFP_cl2 (P881): c = 288,88 ng/µl<br /> | ||
| + | pSB1C3_lITR_CMV_betaglobin_CFP_cl3 (P882): c = 288,36 ng/µl<br /> | ||
| + | |||
| + | <b>Test Digestion:</b> | ||
| + | <br /> | ||
| + | {| border="1" | ||
| + | | <b>components</b> || align="right" |<b>volume P880 - P882 /µl</b> || align="right" |<b>volume P434 /µl </b> | ||
| + | |- | ||
| + | | DNA || align="right" |2 || align="right" |2 | ||
| + | |- | ||
| + | | BSA (10x) || align="right" |1 || align="right" |1 | ||
| + | |- | ||
| + | | Buffer 4 (10x)|| align="right" |1 || align="right" |1 | ||
| + | |- | ||
| + | |Enzyme NgoMIV|| align="right" |0,3|| align="right" |0,3 | ||
| + | |- | ||
| + | |Enzyme AgeI|| align="right" |0,3|| align="right" |0,3 | ||
| + | |- | ||
| + | |H2O|| align="right" |5,4|| align="right" |5,4 | ||
| + | |- | ||
| + | |'''Total volume'''|| align="right" | 10|| align="right" | 10 | ||
| + | |} | ||
| + | |||
| + | <br /> | ||
| + | <b>Gel:</b><br /> | ||
| + | 0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 115 Volt<br /> | ||
| + | |||
| + | |||
| + | <br/> | ||
| + | [[Image:Freiburg10_Test digestion of pSB1C3_lITR_CMV_ß-globin_CFP.jpg|thumb|center|350px|]]<br/> | ||
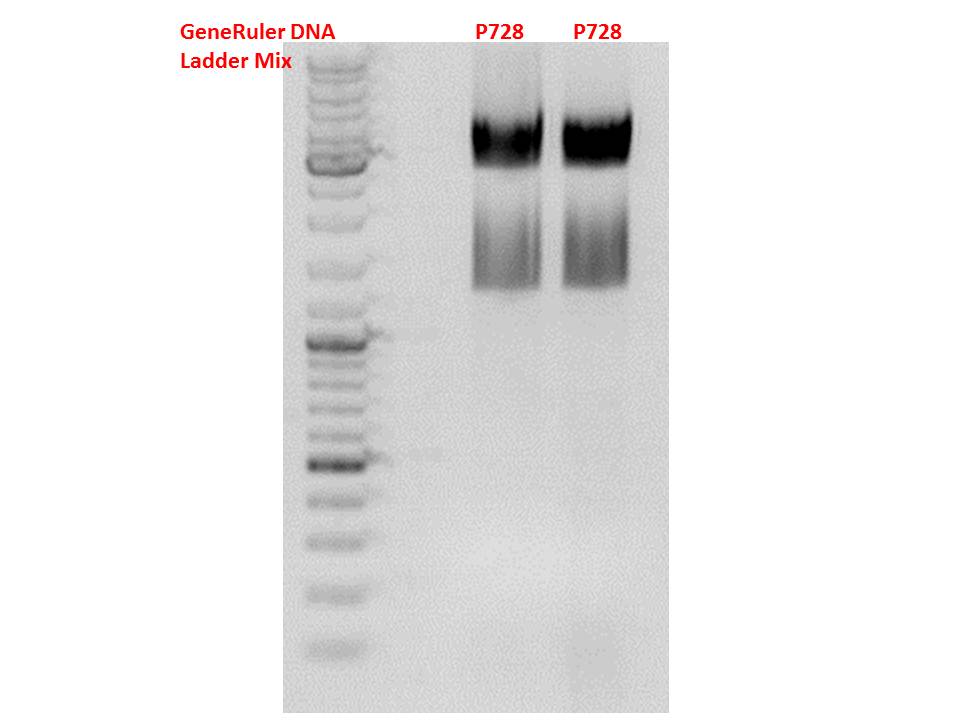
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b> | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Cloning of hGH_rITR into pSB1C3_lITR_CMV_betaglobin_CFP</b></p>==== |
<b>Investigator: Stefan</b><br> | <b>Investigator: Stefan</b><br> | ||
| - | <p style="font-size:13px; color:red;"> | + | <p style="font-size:13px; color:red;">Cloning of our last GOI!</p><br /> |
Vector name:<br /> | Vector name:<br /> | ||
| - | + | pSB1C3_lITR_CMV_betaglobin_CFP cl 1-3 (P880-P882)<br /> | |
| - | + | ||
Insert name:<br /> | Insert name:<br /> | ||
| - | + | pSB1C3_hGH_rITR (P728)<br /> | |
| - | + | ||
<b>Digestion:</b><br /><br /> | <b>Digestion:</b><br /><br /> | ||
| Line 111: | Line 415: | ||
<br /> | <br /> | ||
{| border="1" | {| border="1" | ||
| - | | <b>components</b> || align="right" |<b>volume | + | | <b>components</b> || align="right" |<b>volume P880 - P882 /µl</b> || align="right" |<b>volume P728 /µl </b> |
|- | |- | ||
| - | | DNA || align="right" | | + | | DNA || align="right" |3 || align="right" |8 |
|- | |- | ||
| - | | BSA (10x) | + | | BSA (10x) || align="right" |2 || align="right" |2 |
|- | |- | ||
| - | | Buffer 4 (10x) | + | | Buffer 4 (10x)|| align="right" |2 || align="right" |2 |
|- | |- | ||
| - | |Enzyme | + | |Enzyme PstI|| align="right" |1|| align="right" |1 |
|- | |- | ||
| - | |Enzyme XbaI | + | |Enzyme XbaI|| align="right" |-|| align="right" |1 |
|- | |- | ||
| - | |Enzyme SpeI | + | |Enzyme SpeI|| align="right" |1|| align="right" |- |
|- | |- | ||
| - | |H2O|| align="right" | | + | |H2O|| align="right" |11|| align="right" |4 |
|- | |- | ||
| - | |'''Total volume (e.g. 15,20,25,30 µl)''' | + | |'''Total volume (e.g. 15,20,25,30 µl)'''|| align="right" | 20|| align="right" | 20 |
|} | |} | ||
| Line 136: | Line 440: | ||
<br/> | <br/> | ||
| - | [[Image:Freiburg10 | + | [[Image:Freiburg10 171010.jpg|550px|thumb|center]]<br/> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <p style="font-size:13px; color:red;">Test digestion of all constructs looked alright, therefore, cloning was continued using P881 only.</p><br /> | ||
| + | <br/> | ||
<b>Gel extraction</b>: <br> | <b>Gel extraction</b>: <br> | ||
Was performed according to protocol. | Was performed according to protocol. | ||
| Line 149: | Line 451: | ||
<b>T4 Ligation</b>: <br> | <b>T4 Ligation</b>: <br> | ||
{| border="1" | {| border="1" | ||
| - | |ligation name || align="right" | | + | |ligation name || align="right" |728 + 881 |
|- | |- | ||
| - | |volume of vector | + | |volume of vector || align="right" |2,68 |
|- | |- | ||
| - | |volume of insert | + | |volume of insert|| align="right" |5,32 |
|- | |- | ||
| - | |T4 ligase buffer (10x) | + | |T4 ligase buffer (10x)|| align="right" |1 |
|- | |- | ||
| - | |T4 ligase | + | |T4 ligase || align="right" |1 |
|- | |- | ||
|} | |} | ||
| Line 163: | Line 465: | ||
<br> | <br> | ||
<b>Transformation</b>: <br> | <b>Transformation</b>: <br> | ||
| - | + | Transformation was performed according to standard protocol using BL21 cells. | |
<br/> | <br/> | ||
| - | ====<p style="font-size:15px; background-color:#66bbff;"><b> | + | ====<p style="font-size:15px; background-color:#66bbff;"><b>RT-PCR</b></p>==== |
| - | <b>Investigator: | + | <b>Investigator: Kira</b><br> |
| + | For further experiments, RNA has to be translated into cDNA. The PCR was performed according to the manufacturer protocol. | ||
| - | + | ===<p style="font-size:17px; background-color:#00dd77;">153. labday 18.10.2010</p>=== | |
| + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>quantitative real-time PCR for detection of virus titer</b></p>==== | ||
| + | '''Investigator: Achim <br> | ||
| - | * | + | *qPCR of harvested virus particles to determine the virus titers of our different constructs |
| - | + | *Total number of samples: 58 | |
| - | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Test-digestion of pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_hGH_rITR_SDM-PstI clone 1, 2 and 3 (P877, 878 and 879) </b></p>==== | |
| + | '''Investigator Patrick <br> | ||
| - | + | Check the plasmid for leftITR: | |
| + | 0,5 µl EcoRI, 1 µl BstEII, 7 µl DNA, 2 µl Buffer 4, 2 µl BSA, 7,5 µl H2O, 45 minutes 37°C, 45 minutes 60°C | ||
| + | <br> | ||
| + | <br> | ||
| + | Check the plasmid for hGH_rITR and ... : | ||
| + | 0,5 µl AgeI, 0,5 µl PstI, 3 µl DNA, 1 µl BSA, 1 µl Buffer 4, 4 µl H2O, 70 minutes 37°C | ||
| - | + | Expected results: | |
| + | * leftITR: about 190 bp | ||
| + | * hGH_rITR: about 670 bp | ||
| - | + | <br> | |
| - | < | + | Unfortunately the digestions had to be reapeated because i didnt switch on the current so the samples and especially the 1kb GeneRuler marker diffused. |
| + | <br> | ||
| + | <br> | ||
| + | The second run: see above <br> | ||
| + | [[Image:Freiburg10_1019pat_test1bfertig.jpg]] | ||
| + | <br> | ||
| + | PstI or BstEII seems to work not properly | ||
| - | + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>MTT Assay </b></p>==== | |
| + | '''Investigator Kerstin, Anissa <br> | ||
| + | [[Image:Freiburg10 transductionplan 18.10..jpg]] | ||
| - | + | [[Image:Freiburg10 transductionplan1 18.10..jpg|thumb|center|920px]] | |
| - | + | ||
| + | [[Image:Freiburg10 transductionplan2 18.10..jpg|thumb|center|920px]] | ||
| - | + | '''Results:''' | |
| - | + | ||
| + | [[Image:Freiburg10 Results of MTTAssay 18 10 10 2day 1pic.jpg|thumb|center|920px]] | ||
| + | |||
| + | [[Image:Freiburg10 Results of MTTAssay 18 10 10 2day 3pic.jpg|thumb|center|920px]] | ||
| + | |||
| + | [[Image:Freiburg10 Results of MTTAssay 18 10 10 2day 2pic.jpg|thumb|center|920px]] | ||
| + | |||
| + | ====<p style="font-size:15px; background-color:#66bbFF;"><b>Mini-Prep and test digestion of several constructs</b></p>==== | ||
| + | <b>Investigator: Jessica</b><br /> | ||
| + | |||
| + | Glycerol stocks were prepared:<br> | ||
| + | |||
| + | <ul> | ||
| + | <li>B702 = pSB1C3_lITR_pTERT_ßglobin_CD_hGH_rITR clone 1</li> | ||
| + | <li>B703 = pSB1C3_lITR_pTERT_ßglobin_CD_hGH_rITR clone 2</li> | ||
| + | <li>B704 = pSB1C3_001_VP3 clone 1</li> | ||
| + | <li>B705 = pSB1C3_001_VP3 clone 2</li> | ||
| + | <li>B706 = pSB1C3_001_VP2 clone 1</li> | ||
| + | <li>B707 = pSB1C3_001_VP2 clone 2</li> | ||
| + | <li>B708 = pSB1C3_001_Rep78 clone 1</li> | ||
| + | <li>B709 = pSB1C3_001_Rep78 clone 2</li> | ||
| + | <li>B710 = pSB1C3_001_Rep52 clone 1</li> | ||
| + | <li>B711 = pSB1C3_001_Rep52 clone 2</li> | ||
| + | <li>B712 = pSB1C3_lITR_CMV_ßglobin_CD_hGH_rITR clone 1</li> | ||
| + | <li>B713 = pSB1C3_lITR_CMV_ßglobin_CD_hGH_rITR clone 2</li> | ||
| + | <li>B714 = pSB1C3_001_VP1 clone 1</li> | ||
| + | <li>B715 = pSB1C3_001_VP1 clone 2</li> | ||
| + | |||
| + | </ul><br> | ||
| + | |||
| + | Mini-Prep was performed according to standard protocol:<br> | ||
| + | |||
| + | <ul> | ||
| + | <li>P886 = pSB1C3_lITR_pTERT_ßglobin_CD_hGH_rITR clone 1 c= 232,2ng/µl</li> | ||
| + | <li>P887 = pSB1C3_lITR_pTERT_ßglobin_CD_hGH_rITR clone 2 c= 186,2ng/µl</li> | ||
| + | <li>P888 = pSB1C3_001_VP3 clone 1 c= 300,8ng/µl</li> | ||
| + | <li>P889 = pSB1C3_001_VP3 clone 2 c= 284,4ng/µl</li> | ||
| + | <li>P890 = pSB1C3_001_VP2 clone 1 c= 298,3ng/µl</li> | ||
| + | <li>P891 = pSB1C3_001_VP2 clone 2 c= 299,9ng/µl</li> | ||
| + | <li>P892 = pSB1C3_001_Rep78 clone 1 c= 143,9ng/µl</li> | ||
| + | <li>P893 = pSB1C3_001_Rep78 clone 2 c= 163,4ng/µl</li> | ||
| + | <li>P894 = pSB1C3_001_Rep52 clone 1 c= 166,6ng/µl</li> | ||
| + | <li>P895 = pSB1C3_001_Rep52 clone 2 c= 181,6ng/µl</li> | ||
| + | <li>P896 = pSB1C3_lITR_CMV_ßglobin_CD_hGH_rITR clone 1 c= 250,5ng/µl</li> | ||
| + | <li>P897 = pSB1C3_lITR_CMV_ßglobin_CD_hGH_rITR clone 2 c= 173,4ng/µl</li> | ||
| + | <li>P898 = pSB1C3_001_VP1 clone 1 c= 272,7ng/µl</li> | ||
| + | <li>P899 = pSB1C3_001_VP1 clone 2 c= 294,8ng/µl</li> | ||
| + | <li>P900 = pSB1C3_hGH_rITR (from B160) c= 136,7ng/µl</li> | ||
| + | </ul> | ||
| + | |||
| + | Test digestion:<br> | ||
| + | {| border="1" | ||
| + | | align="left" | '''Components''' ||align="left"| <b>P886,887,892,893,894,895,896,897 / µl</b>||align="left"| <b>P888,889,890,891898,899 / µl</b> | ||
| + | |- | ||
| + | | align="left" | DNA ||align="left"| 1,5||align="left"| 1,5 | ||
| + | |- | ||
| + | | align="left" | Buffer ||align="left"| (4) 1||align="left"| (2) 1 | ||
| + | |- | ||
| + | | align="left" | BSA (10x) ||align="left"| 1||align="left"| 1 | ||
| + | |- | ||
| + | | align="left" | NgoMIV ||align="left"| 0,4 ||align="left"| - | ||
| + | |- | ||
| + | | align="left" | XbaI ||align="left"| 0,4 ||align="left"| - | ||
| + | |- | ||
| + | | align="left" | PstI ||align="left"| - ||align="left"| 0,6 | ||
| + | |- | ||
| + | | align="left" | XcmI ||align="left"| - ||align="left"| 0,4 | ||
| + | |- | ||
| + | | align="left" | H<sub>2</sub>O ||align="left"| 4,5||align="left"| 4,5 | ||
| + | |- | ||
| + | | align="left" | '''Total volume''' ||align="left"| <b>10</b> ||align="left"| <b>10</b> | ||
| + | |} | ||
| + | |||
| + | Gel:<br> | ||
| + | 1,0g agarose, 100 ml TAE (1%), 6 µl GELRED, Volt, running time minutes<br> | ||
| + | |||
| + | [[Image:Freiburg10 test digestion 886-899.jpg|thumb|center|800px]] | ||
| + | <p style="color:#66bbff;"><i>Comment</i>: Rep 52/78 will be checked,pSB1C3_lITR_CMV_ßglobin_CD_hGH_rITR and pSB1C3_lITR_pTERT_ßglobin_CD_hGH_rITR will be sequenced</p> | ||
| + | |||
| + | ====<p style="font-size:15px; background-color:#66bbff;"><b>PCR of mGMK and SR39</b></p>==== | ||
| + | <b>Investigator: Anna</b><br> | ||
| + | |||
| + | |||
| + | Plasmids:<br /> | ||
| + | pSB1C3_mGMK_TK30_SDM-PstI clone 2(P804)<br /> | ||
| + | pSB1C3_mGMK_sr39 clone 1(P860)<br /> | ||
| + | |||
| + | Oligos:<br /> | ||
| + | O193: pTK30_for<br /> | ||
| + | O81: pmgmk_tk30_suffix_RFC25_rev | ||
| + | <br /> | ||
| + | |||
| + | |||
| + | <b>PCR Mix:</b> | ||
| + | |||
| + | {| border="1" | ||
| + | | <b>Components</b> || align="right" |<b>Volume /µl</b> | ||
| + | |- | ||
| + | | Phusion Buffer || align="right" |10 | ||
| + | |- | ||
| + | | dNTP || align="right" |1 | ||
| + | |- | ||
| + | |Primer_for|| align="right" |2,5 | ||
| + | |- | ||
| + | |Primer_rev|| align="right" |2,5 | ||
| + | |- | ||
| + | |DNA template|| align="right" |1 | ||
| + | |- | ||
| + | |H2O|| align="right" |32,5 | ||
| + | |- | ||
| + | |'''Total volume'''|| align="right" | 50 | ||
| + | |} | ||
| + | <br /> | ||
| + | <b>PCR Program:</b> | ||
| + | |||
| + | {| border="1" | ||
| + | |Cycles||Temperature||Time | ||
| + | |- | ||
| + | |||98°C||60 sec | ||
| + | |- | ||
| + | |||98°C||15 sec | ||
| + | |- | ||
| + | |8x||52°C||25 sec | ||
| + | |- | ||
| + | |||72°C||25 sec | ||
| + | |- | ||
| + | |||98°C||15 sec | ||
| + | |- | ||
| + | |17x||67°C||25 sec | ||
| + | |- | ||
| + | |||72°C||25 sec | ||
| + | |- | ||
| + | |1x||72°C||5 min | ||
| + | |- | ||
| + | |Hold 4°C | ||
| + | |} | ||
| + | |||
| + | <br /> | ||
| + | <b>Gel:</b><br /> | ||
| + | 0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 115 Volt<br /> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br/> | <br/> | ||
| - | + | [[Image:|550px|]]<br/> | |
| - | + | ||
| - | + | ===<p style="font-size:17px; background-color:#00dd77;">154. labday 19.10.2010</p>=== | |
| - | + | ||
| - | + | ====<p style="font-size:15px; background-color:#66bbff;">Midi-Prep</p>==== | |
| - | + | ||
| - | + | '''Investigator: Chris W. <br>''' | |
| - | + | <p style="font-size:13px; color:#003399;"> Midi-Prep of:</p><br/> | |
| + | pSB1C3_lITR_CMV_betaglobin_mVenus_hGH_rITR clone1 =P901 =B200<br/> | ||
| + | pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_hGH_rITR_SDM-PstI clone 2 =P902 =B697<br/> | ||
| + | |||
| + | |||
| + | |||
| + | |||
<br/> | <br/> | ||
| - | + | The Midi-Preps were performed according to the standard protocol yielding the following concentrations: | |
| - | + | ||
| - | + | ||
| - | ===<p style="font-size: | + | {| border="1" |
| + | | plasmid-no. || align="right" |P901|| align="right" |P902 | ||
| + | |- | ||
| + | | concentration (ng/µl)|| align="right" |1563,63 || align="right" |1348,26 | ||
| + | |} | ||
| + | <br> | ||
| + | <br/> | ||
| + | ====<p style="font-size:15px; background-color:#66bbff;"><b>Continuation of PCR of mGMK and SR39</b></p>==== | ||
| + | <b>Investigator: Jessica</b><br> | ||
| - | + | * vector (P320) and PCR product was digested<br> | |
| - | ===< | + | {| border="1" |
| + | | align="left" | '''Components''' ||align="left"| <b>P320 / µl</b>||align="left"| <b>PCR product P804 and P860 / µl</b> | ||
| + | |- | ||
| + | | align="left" | DNA ||align="left"| 1,5||align="left"| 20 | ||
| + | |- | ||
| + | | align="left" | Buffer ||align="left"| 2||align="left"| 3 | ||
| + | |- | ||
| + | | align="left" | BSA (10x) ||align="left"| 2||align="left"| 3 | ||
| + | |- | ||
| + | | align="left" | AgeI ||align="left"| 1 ||align="left"| 1,5 | ||
| + | |- | ||
| + | | align="left" | XbaI ||align="left"| 1 ||align="left"| 1,5 | ||
| + | |- | ||
| - | ===< | + | | align="left" | H<sub>2</sub>O ||align="left"| 14,5||align="left"| 1 |
| + | |- | ||
| + | | align="left" | '''Total volume''' ||align="left"| <b>20</b> ||align="left"| <b>30</b> | ||
| + | |} | ||
| + | [[Image:Freiburg10 pcr mGMK and SR39.jpg|thumb|center|800px]] | ||
| + | <br> | ||
| + | <br> | ||
| + | '''Ligation''' | ||
| + | * P320 c= 5,08 ng/µl | ||
| + | * P804 c= 11,73 ng/µl | ||
| + | * P860 c= 26,57 ng/µl | ||
| + | <br> | ||
| + | * P320 + P804: 4,93µl : 3,07µl | ||
| + | * P320 + P860: 6,28µl : 1,72µl | ||
| + | <br> | ||
| + | '''Transformation with BL21 and Cm''' | ||
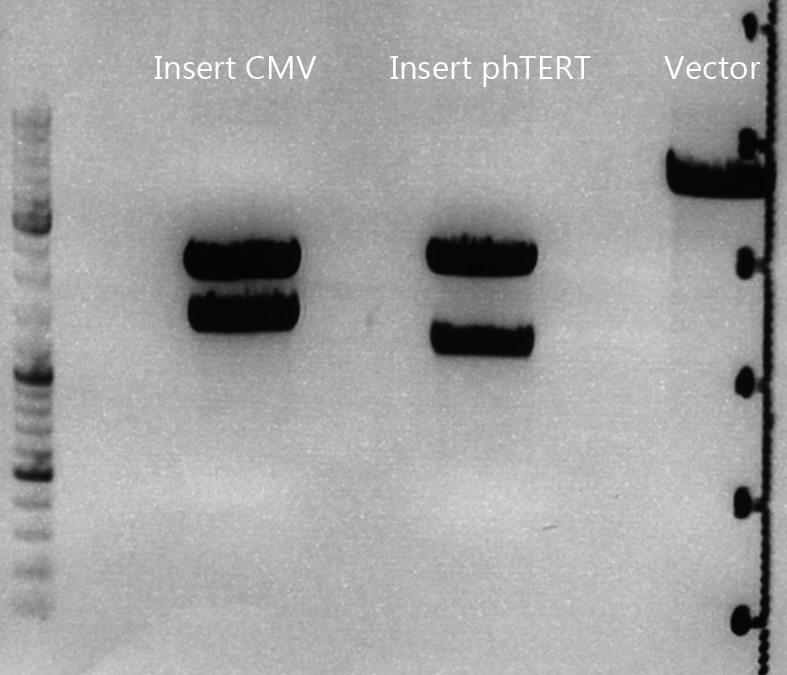
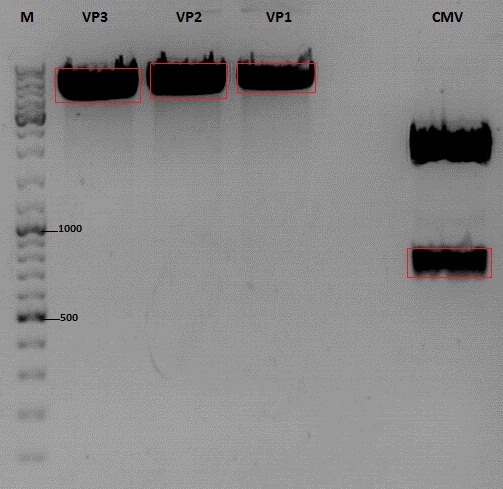
| - | ===<p style="font-size: | + | ====<p style="font-size:15px; background-color:#66bbff;">Cloning of CMV into pSB1C3_001_VP1, pSB1C3_001_VP2 and pSB1C3_001_VP3</p>==== |
| - | + | '''Investigator: Kerstin, Anna''' | |
| + | <br> | ||
| - | ===< | + | '''Plasmids:''' |
| + | *P888: pSB1C3_001_VP3, c = 300,8 ng/µl | ||
| + | *P890: pSB1C3_001_VP2, c = 298,3 ng/µl | ||
| + | *P898: pSB1C3_001_VP1, c = 272,7 ng/µl | ||
| + | *P727: pSB1C3_001_CMV, c = 225,5 ng/µl | ||
| + | <br> | ||
| + | '''Digestion:''' | ||
| + | {| border="1" | ||
| + | | <b>components</b> || align="right" |<b>VP3</b> || align="right" |<b>VP2</b> || align="right" |<b>VP1</b>|| align="right" |<b>CMV</b> | ||
| + | |- | ||
| + | | DNA || align="right" |4 || align="right" |4|| align="right" |4|| align="right" |8 | ||
| + | |- | ||
| + | | BSA (10x) || align="right" |2 || align="right" |2|| align="right" |2|| align="right" |2 | ||
| + | |- | ||
| + | | Buffer 4 (10x)|| align="right" |2 || align="right" |2 || align="right" |2|| align="right" |2 | ||
| + | |- | ||
| + | |Enzyme EcoI|| align="right" |1|| align="right" |1 || align="right" |1|| align="right" |1 | ||
| + | |- | ||
| + | |Enzyme XbaI|| align="right" |1|| align="right" |1 || align="right" |1 || align="right" |- | ||
| + | |- | ||
| + | |Enzyme SpeI|| align="right" |-|| align="right" |- || align="right" |-|| align="right" |1 | ||
| + | |- | ||
| + | |H2O|| align="right" |10|| align="right" |10 || align="right" |10 || align="right" |10 | ||
| + | |- | ||
| + | |'''Total '''|| align="right" | 20|| align="right" | 20|| align="right" | 20 | ||
| + | |} | ||
| + | <br> | ||
| + | *Digestion: 2h @ 37°C | ||
| + | <br> | ||
| + | |||
| + | '''Gel:''' | ||
| + | *1% agarose gel, 1 µl Gelred, run for 45 min | ||
| + | |||
| + | [[Image:Freiburg10_Cloning of CMV into pSB1C3_001_VP1-3.jpg|400px]] | ||
| + | <br> | ||
| + | |||
| + | '''Gel extraction''' | ||
| + | |||
| + | {| border="1" | ||
| + | |sample name || align="right" |<b>VP3</b>|| align="right" |<b>VP2</b>|| align="right" |<b>VP1</b>|| align="right" |<b>CMV</b> | ||
| + | |- | ||
| + | |nanodrop concentrations || align="right" |44,36|| align="right" |32,33|| align="right" |22,73|| align="right" |18,5 | ||
| + | |- | ||
| + | |expected fragment size|| align="right" |4100|| align="right" |4000|| align="right" |3700|| align="right" |650 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | <br> | ||
| + | '''Ligation:''' | ||
| + | {| border="1" | ||
| + | |ligation name || align="right" |<b>VP3 + CMV</b>|| align="right" |<b>VP2 + CMV</b>|| align="right" |<b>VP1 + CMV</b> | ||
| + | |- | ||
| + | |volume of vector || align="right" |5,7|| align="right" |4,3|| align="right" |5 | ||
| + | |- | ||
| + | |volume of insert|| align="right" |3,3|| align="right" |3,7|| align="right" |3 | ||
| + | |- | ||
| + | |T4 ligase buffer (10x)|| align="right" |1|| align="right" |1|| align="right" |1 | ||
| + | |- | ||
| + | |T4 ligase || align="right" |1|| align="right" |1|| align="right" |1 | ||
| + | |- | ||
| + | |} | ||
| + | *Ligation @ RT for 30 min | ||
| + | |||
| + | '''Trafo:''' | ||
| + | <br> | ||
| + | |||
| + | Was done following the standard protocol using BL21 cells. | ||
| + | |||
| + | |||
| + | <br> | ||
===<p style="font-size:17px; background-color:#00dd77;">155. labday 20.10.2010</p>=== | ===<p style="font-size:17px; background-color:#00dd77;">155. labday 20.10.2010</p>=== | ||
| + | |||
| + | ====<p style="font-size:15px; background-color:#66bbff;">Midi-Prep</p>==== | ||
| + | |||
| + | '''Investigator: Chris W. <br>''' | ||
| + | <p style="font-size:13px; color:#003399;"> Midi-Prep of:</p><br/> | ||
| + | pSB1C3_lITR_pTERT_ßglobin_CD_hGH_rITR clone 1 =P903 =B702<br/> | ||
| + | pSB1C3_lITR_CMV_ßglobin_CD_hGH_rITR clone 1 =P904 =B712<br/> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <br/> | ||
| + | The Midi-Preps were performed according to the standard protocol yielding the following concentrations: | ||
| + | |||
| + | {| border="1" | ||
| + | | plasmid-no. || align="right" |P903|| align="right" |P904 | ||
| + | |- | ||
| + | | concentration (ng/µl)|| align="right" |832,63 || align="right" |1174,49 | ||
| + | |} | ||
| + | <br> | ||
| + | <br/> | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">156. labday 21.10.2010</p>=== | ||
| + | ====<p style="font-size:15px; background-color:#66bbff;">ÄKTA Chromatography and Ultrafiltration of virus particles </p>==== | ||
| + | |||
| + | '''Investigator: Hanna <br>''' | ||
| + | <b>ÄKTA chromatography</b> with VP1up_NLS_mVenus_VP2/3 containing virus particles was conducted. Fraction 5 - 10 delivered highest protein concentrations. <br/> | ||
| + | {| border="1" | ||
| + | |<b>Sample</b> || align="right" |<b>A(260 nm)</b> || align="right" |<b>A(280 nm)</b>|| align="right" |<b>A(515 nm) (YFP)</b> | ||
| + | |- | ||
| + | | 5 || align="right" |0.032 || align="right" |0.027|| align="right" |0.003 | ||
| + | |- | ||
| + | | 6 || align="right" |0.019 || align="right" |0.019|| align="right" |0.003 | ||
| + | |- | ||
| + | | 7|| align="right" |0.075 || align="right" |0.09 || align="right" |0.01 | ||
| + | |- | ||
| + | |8|| align="right" |0.054|| align="right" |0.075 || align="right" |0.007 | ||
| + | |- | ||
| + | |10|| align="right" |-0.008|| align="right" |-0.008 || align="right" |0.005 | ||
| + | |} | ||
| + | |||
| + | <br/> | ||
| + | A further attempt was conducted which included digestion with Benzonase (1 hour) prior to ÄKTA chromatography. Following protein concentrations were obtained: <br/> | ||
| + | {| border="1" | ||
| + | |<b>Sample</b> || align="right" |<b>A(260 nm)</b> || align="right" |<b>A(280 nm)</b>|| align="right" |<b>A(515 nm) (YFP)</b> | ||
| + | |- | ||
| + | | 5 || align="right" |0.005|| align="right" |0.007|| align="right" |0.003 | ||
| + | |- | ||
| + | | 6 || align="right" |0.047|| align="right" |0.041|| align="right" |0.006 | ||
| + | |- | ||
| + | | 7|| align="right" |0.151|| align="right" |0.153|| align="right" |0.01 | ||
| + | |- | ||
| + | |8|| align="right" |0.172|| align="right" |0.2|| align="right" |0.009 | ||
| + | |- | ||
| + | |9|| align="right" |0.098|| align="right" |0.128|| align="right" |0.009 | ||
| + | |- | ||
| + | |10|| align="right" |0.053|| align="right" |0.074|| align="right" |0.005 | ||
| + | |} | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <b>Ultrafiltration</b> <br/> | ||
| + | Ultrafiltration of CFP_MiddleLinker_VP2/3 containing virus particles and 587-BAP virus particles were concentrated via Vivaspin-Ultrafiltration: <br/> | ||
| + | *20 mL virus containing cell culture supernatant was added to GE Vivaspin 20 filter and centrifuged with 4000 g at 15°C until 750 - 1000 µL was left- | ||
| + | * 5 mL Bis-Trus buffer (pH 6) was added and centrifuged again with 4000 g at 15°C (washing). | ||
| + | * This step was repeated 3 more times. | ||
| + | * Membrane was carefully resuspended and cleared. Suspension was transfered to low-binding eppi and centrifuged with 10000 g for 10 minutes at 15°C. | ||
| + | * Supernatant was transfered to new low-binding eppi and again centrifuged with 10000 g for 10 minutes at 15°C. | ||
| + | * Supernatant was transfered to new low-binding eppi and stored at 4°C over night. <b>To do:</b> ÄKTA chromatography. <br/> | ||
| + | |||
| + | |||
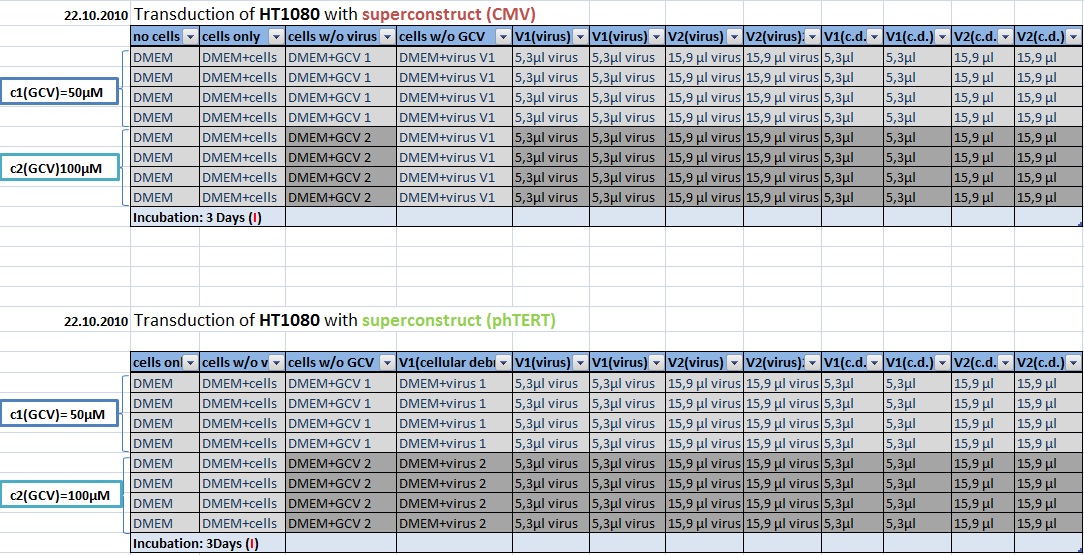
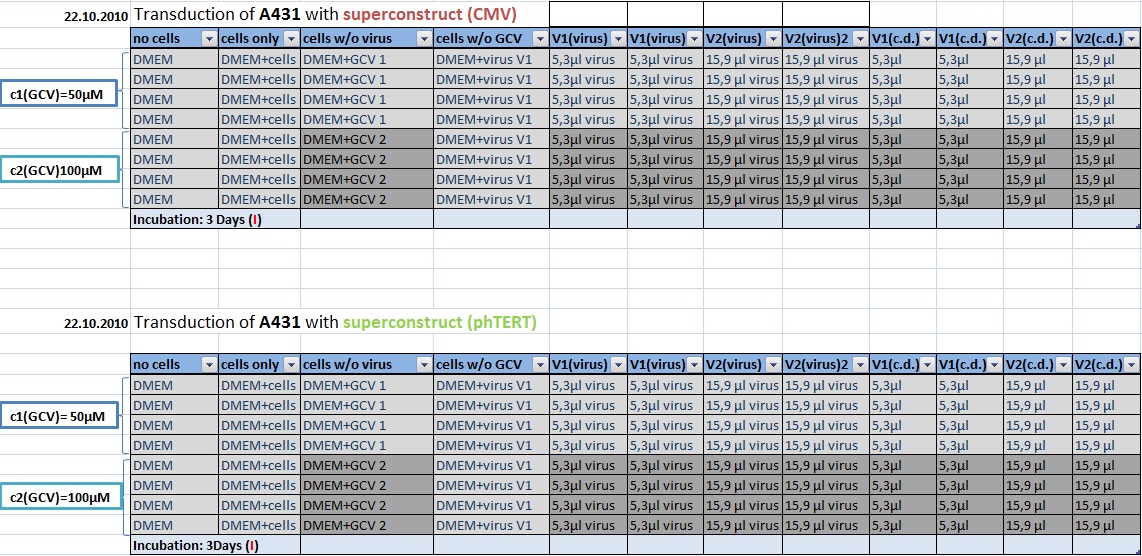
| + | ====<p style="font-size:15px; background-color:#66bbff;">MTT Assay: Testing Superconstructs </p>==== | ||
| + | '''Investigator: Anissa, Kerstin <br>''' | ||
| + | |||
| + | [[Image:Freiburg10 Transductionplan1 21.10.2010.jpg|700px]] | ||
| + | [[Image:Freiburg10 Transductionplan2 21.10.2010.jpg|700px]] | ||
| + | [[Image:Freiburg10 Transductionplan3 21.10.2010.jpg|700px]] | ||
| + | [[Image:Freiburg10 Transductionplan4 21.10.2010.jpg|700px]] | ||
| + | [[Image:Freiburg10 Transductionplan5 21.10.2010.jpg|700px]] | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">157. labday 22.10.2010</p>=== | ||
| + | ====<p style="font-size:15px; background-color:#66bbff;">SDS PAGE and Coomassie staining</p>==== | ||
| + | |||
| + | '''Investigator: Hanna <br>''' | ||
| + | <br/> | ||
| + | Prior to performing Western Blot we decided to investigate running behaviour of different samples. <br/> | ||
| + | * 1. Cell debris (control) | ||
| + | * 2. Cell debris containing virus particles | ||
| + | * 3. Concentrated virus stock (containing CFP_MiddleLinker_VP2/3) | ||
| + | * 4. ÄKTA purified virus stock: Fraction 6 | ||
| + | * 5. ÄKTA purified virus stock: Fraction 7 | ||
| + | * 6. Benzonase treated, ÄKTA purified virus stock: Fraction 7 | ||
| + | * 7. Benzonase treated, ÄKTA purified virus stock: Fraction 8 | ||
| + | |||
| + | <br/> | ||
| + | 5 µL Laemmli buffer was added to 20 µL sample. Samples were incubated at 95°C for 8 minutes and loaded onto a SDS gel (10 %). SDS PAGE was performed at 90 V (collection gel) resp. 120 V (separation gel). <br/> | ||
| + | Gel was put into Coomassie dye, heated for 30 seconds in microwave and incubated for 1 hours shaking. <br/> | ||
| + | Gel was decolorized in acetic acid (20%). <br/> | ||
| + | Loading plan: | ||
| + | {| border="1" | ||
| + | |<b>Marker</b> || align="right" |<b>Concentrated Stock</b> || align="right" |<b>ÄKTA 6</b>|| align="right" |<b>ÄKTA 7</b>|| align="right"|<b>Benzonase/ÄKTA 7</b>|| align="right" |<b>Benzonase/ÄKTA 8</b>|| align="right"|<b> - - - </b>|| align="right" |<b> - - - </b>|| align="right"|<b>Cell debris</b>|| align="right" |<b>Cell debris + Virus</b> | ||
| + | |} | ||
| + | <br/> | ||
| + | [[Image:Freiburg10 SDSVersuch2.jpg|500px|center]] | ||
| + | <br/> | ||
| + | Gel picture shows that concentration works :) <br/> | ||
| + | In addition to that one can see that the BSA bands disappear after ÄKTA chromatography. <br/> | ||
| + | <b>Next step:</b> Western Blot of ÄKTA purified virus stocks. <br/> | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">158. labday 23.10.2010</p>=== | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">159. labday 24.10.2010</p>=== | ||
| + | |||
| + | ====<p style="font-size:15px; background-color:#66bbff;">Midi-Prep</p>==== | ||
| + | |||
| + | '''Investigator: Chris W. <br>''' | ||
| + | <p style="font-size:13px; color:#003399;"> Midi-Prep of:</p><br/> | ||
| + | pSB1C3_001_RC_IRCK_VP2-ko_HSPG-ko_P5tataless cl1 =P966 =B523<br/> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <br/> | ||
| + | The Midi-Prep were performed according to the standard protocol yielding the following concentration: | ||
| + | |||
| + | {| border="1" | ||
| + | | plasmid-no. || align="right" |P966 | ||
| + | |- | ||
| + | | concentration (ng/µl)|| align="right" |310,69 | ||
| + | |} | ||
| + | <br> | ||
| + | <br/> | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">160. labday 25.10.2010</p>=== | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">161. labday 26.10.2010</p>=== | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">162. labday 27.10.2010</p>=== | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">163. labday 28.10.2010</p>=== | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">164. labday 29.10.2010</p>=== | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">165. labday 30.10.2010</p>=== | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">166. labday 31.10.2010</p>=== | ||
<!-- End of page--> | <!-- End of page--> | ||
| - | <center>[https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/ | + | <center>[https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/November '''=> Go to Labjournal November (labday 167 - 170 )''']</center><br> |
<html> | <html> | ||
</div> | </div> | ||
</html> | </html> | ||
Latest revision as of 21:33, 27 October 2010
- March (labday 1)
- April (labday 2 - 5)
- May (labday 6 - 17)
- June (labday 18 - 45)
- July (labday 46 - 75)
- August part 1 (labday 76 - 92)
- August part 2 (labday 93 - 106)
- September part 1 (labday 107 - 123)
- September part 2 (labday 124 - 135)
- October part 1 (labday 136 - 149 )
- October part 2 (labday 150 - 166 )
- November (labday 167 - 170 )
- Cellculture
150. labday 15.10.2010
Cloning CFP (from P666: PSB1C3_CFP) into pSB1C3_leftITR_CMV_beta-globin (P729)
Investigator Patrick
Digestions, 2 h 10 minutes, 37 °C:
- P666: 5 µl DNA, 2 µl BSA, 2 µl Buffer 4 (10x), 1 µl Xba, 1 µl PstI, 9 µl H2O
- P729: 4 µl DNA, 2 µl BSA, 2 µl Buffer 4 (10x), 1 µl SpeI, 1 µl PstI, 10 µl H2O
Expected results for the 1% agarose gel:
- P666: about 2100 and 750 bp
- P729: about 3300 and 20 bp
The gelextraction ...
P728: 11,8 ng/µl
P822: 34,6 ng/µl
... and following ligation (2,5 µl Insert, 5,5 µl vector, 1 µl T4 DNA Ligase, 1 µl T4 DNA Ligase Buffer (10x), 40 minutes, RT) were performed according to the standard protocol. After the transformation (with XL1B) the cells were plated and put into the 37°C room.
Two additional transformations were performed with ligations from Volker labeled: "ligation viral brick 453 empty" & "viral brick 587 empty".
The following day the plates were checked for clones. Unfortunately there grew no clones on these two plates contrary to my plate with a a lot of clones.
Midi-Prep
Investigator: Chris W.
Midi-Prep of:
pSB1C3_001_RC_IRCK_P5tataless clone 1 =P866 =B516
pSB1C3_001_CMV_VP123_587-KO_Z34C_spacer clone2 =P867 =B526
pSB1C3_001_CMV_VP123_587-KO_Z34C clone2 =P868 =B529
pSB1C3_CMV_Zegfr:1907_MiddleLinker_VP2/3_587-KO_BAP clone 1 =P869 =B680
pSB1C3_CMV_Zegfr:1907_MiddleLinker_VP2/3_587-KO_6xHis clone 1 =P870 =B200
The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| plasmid-no. | P866 | P867 | P868 | P869 | P870 |
| concentration (ng/µl) | 899,80 | 954,46 | 406,97 | 1642,76 | 1585,12 |
mini prep of several constructs
Investigator: Kira
c(rep52_1)=299,04 ng/ul
c(rep52_2)=290,07 ng/ul
c(rep78_1)=142,32 ng/ul
c(rep78_2)=175,36 ng/ul
Cell culture
Investigator: Kira
The cells are still alive. Medium was exchanged.--> RNA will be harvested tomorrow
Mini-Prep and test digestion of pSB1C3_CD_SDM-PstI_hGH_rITR
Investigator: Stefan
Glycerol stocks were prepared:
- B694 = pSB1C3_CD_SDM-PstI_hGH_rITR clone 1
- B695 = pSB1C3_CD_SDM-PstI_hGH_rITR clone 2
Mini-Prep was performed according to standard protocol:
- P875 = pSB1C3_CD_SDM-PstI_hGH_rITR clone 1 c = 73,1 ng/µl
- P876 = pSB1C3_CD_SDM-PstI_hGH_rITR clone 2 c = 78,3 ng/µl
Test digestion:
| Components | P875 + P876 / µl |
| DNA | 4 |
| Buffer 4 | 1 |
| BSA (10x) | 1 |
| XbaI | 0,3 |
| AgeI | 0,3 |
| H2O | 3,4 |
| Total volume | 10 |
Gel:
0,5g agarose, 50 ml TAE (1%), 3 µl GELRED, 115 Volt, running time ~50 minutes
Comment: Test digestion looks allright, cloning will be continued using P876.
151. labday 16.10.2010
Biobrick assembly: pSB1C3_lITR_CMV_ß-globin_CD_hGH_rITR and pSB1C3_lITR_phTERT_ß-globin_CD_hGH_rITR
Investigator: Achim
Plasmids:
- P729: pSB1C3_lITR_CMV_ß-Globin
- c= 243.4 ng/µl
- P730: pSB1C3_lITR_pHTERT_ß-Globin
- c= 81.1 ng/µl
- P876: pSB1C3_CD_SDM-PstI_hGH_rITR
- c= 78.3 ng/µl
Digestion:
| components | I1 (P729) | I2 (P730) | V (P876) |
| DNA | 6 | 14 | 14 |
| BSA (10x) | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 |
| Enzyme EcoI | 1 | 1 | 1 |
| Enzyme XbaI | - | - | 1 |
| Enzyme SpeI | 1 | 1 | - |
| H2O | 8 | - | - |
| Total | 20 | 20 | 20 |
Digestion: 2h, 37°C
Prep. gel:
- 0,8%, run for 45 min
- Corresponding bands were cut out
Gel ex.
- Nanodrop concentrations:
- I1: 37.54 ng/µl
- I2: 25.38 ng/µl
- V: 26.47 ng/µl
Ligation:
| ligation name | I1 + V | I2 + V |
| volume of vector | 4.69 | 4.23 |
| volume of insert | 3.31 | 3.77 |
| T4 ligase buffer (10x) | 1 | 1 |
| T4 ligase | 1 | 1 |
- Ligation @ RT for 40 min
Trafo:
- Done by Kira
Mini-prep of mutual pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_hGH_rITR_SDM-PstI
Investigator Patrick
Yielded concentrations & given numbers:
- pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_hGH_rITR_SDM-PstI clone 1: 208,4 ng/µl , P877 / B696
- pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_hGH_rITR_SDM-PstI clone 2: 251,6 ng/µl , P878 / B696
- pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_hGH_rITR_SDM-PstI clone 3: 212,6 ng/µl , P889 / B696
Test-digestion: 0,5 µl SpeI, 0,5 µl PstI, 3 µl DNA, 1 µl Buffer 4, 1 µl BSA, 4 µl H2O, 40 minutes, 37°C
Expected results: fragments with about 650 and 4900 bp
Obviously, this test digestion has to be repeated.
Preparations for tomorrow:
- Mini-prep of 3 mutual pSB1C3_leftITR_CMV_beta-globin_CFP clones (have to be picked from the plate)
- Midi-prep of pHelper
- Midi-prep of B689:pSB1C3_lITR_CMV_betaglobin_mGMK_TK30_SDM-PstI_hGH_rITR clone 2
Biobrick assembly of Rep78 and Rep52
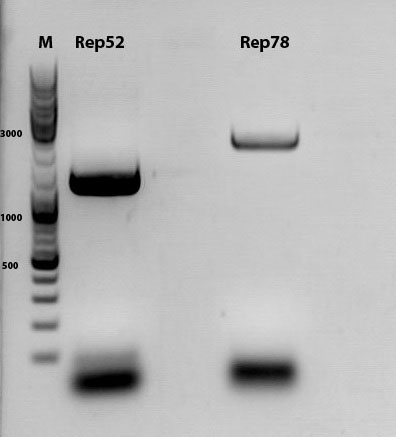
Investigator: Kira
Comment: After replacing the mutated Rep parts by the ordered Rep parts, PCR amplification has to be done in order to produce a biobrick.
PCR program:
c(Rep52)=299 ng/ul
c(Rep78)=175 ng/ul
Rep52: praefix 094 & suffix 097
Rep78: praefix 093 & suffix 097
| components | volume in µl |
| 5x Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| primer_for (1:10 dilution) | 2,5 |
| primer_rev (1:10 dilution) | 2,5 |
| DNA template (1:100) | 0,5 |
| DMSO | 0,5 |
| Phusion polymerase | 0,5 |
| H2O | 32,5 |
| Total volume (e.g. 50 µl) | 50 |
| Cycles | Temperature | Time |
| 98°C | 30 sec | |
| 10x | 98°C | 15 sec |
| 63°C | 25 sec | |
| 72°C | 32 sec | |
| 20x | 98°C | 15 sec |
| 66°C | 25 sec | |
| 72°C | 32 sec | |
| 1x | 72°C | 5 min |
| Hold 4°C |
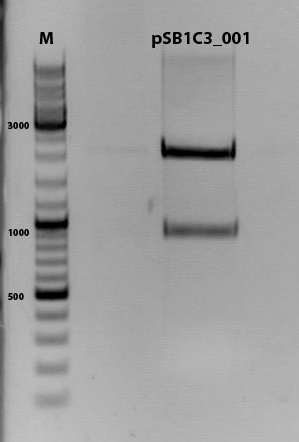
Digestion of plasmid backbone:
pSB1C3_001 is used as backbone
| Components | <b>vector Volume/µL |
| DNA | 3,5 µl |
| BSA (10x) | 2 µl |
| Buffer no. 4 (10x) | 2,0 µl |
| Enzyme 1 EcoRI-HF | 0,5 µl |
| Enzyme 2 SpeI | 1,0 µl |
| H2O | 15 µl |
| Total volume | 25 |
incubation @ 37 C for approx. 2 h
1% agarose gel
Digestion of PCR product:
| Components | PCR product Volume/µL |
| DNA | 35,0 µl |
| BSA (100x) | 0,45 µl |
| Buffer no. 4 | 4,5 µl |
| Enzyme 1 EcoRI-HF | 1,5 µl |
| Enzyme 2 SpeI | 2,0 µl |
| H2O | 1,5 µl |
| Total volume | 45 |
incubation @ 37 C for approx. 2 h
T4 ligation for 40 min
Transformation according to the standard protocol
RNA harvesting
Investigator: Kira
After transfection, the cells were incubated for 48 hours. Today, the cells will be harvested and RNA extracted, in order to perform RT-PCR and an additional PCR for evaluation of promoter activity.
The transfected cells were trypsinised and centrifuged for 2 min. The supernatant was discarded and pellet washed 2x with PBS. RNeasy Kit [Qiagen] was used for RNA extraction according to the manufacturer protocol.
c(CMV)= 335,69 ng/ul
c(P40)= 857,92 ng/ul
c(AAV_RC)= 760,21 ng/ul
152. labday 17.10.2010
Test digestion of pSB1C3_lITR_CMV_ß-globin_CFP
Investigator: Anna
Vector name:
pSB1C3_lITR_CMV_betaglobin_CFP_cl1 (P880): c = 452,67 ng/µl
pSB1C3_lITR_CMV_betaglobin_CFP_cl2 (P881): c = 288,88 ng/µl
pSB1C3_lITR_CMV_betaglobin_CFP_cl3 (P882): c = 288,36 ng/µl
Test Digestion:
| components | volume P880 - P882 /µl | volume P434 /µl |
| DNA | 2 | 2 |
| BSA (10x) | 1 | 1 |
| Buffer 4 (10x) | 1 | 1 |
| Enzyme NgoMIV | 0,3 | 0,3 |
| Enzyme AgeI | 0,3 | 0,3 |
| H2O | 5,4 | 5,4 |
| Total volume | 10 | 10 |
Gel:
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 115 Volt
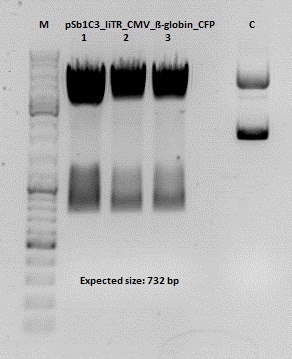
Cloning of hGH_rITR into pSB1C3_lITR_CMV_betaglobin_CFP
Investigator: Stefan
Cloning of our last GOI!
Vector name:
pSB1C3_lITR_CMV_betaglobin_CFP cl 1-3 (P880-P882)
Insert name:
pSB1C3_hGH_rITR (P728)
Digestion:
| components | volume P880 - P882 /µl | volume P728 /µl |
| DNA | 3 | 8 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme PstI | 1 | 1 |
| Enzyme XbaI | - | 1 |
| Enzyme SpeI | 1 | - |
| H2O | 11 | 4 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 |
Gel:
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 115 Volt
Test digestion of all constructs looked alright, therefore, cloning was continued using P881 only.
Gel extraction:
Was performed according to protocol.
T4 Ligation:
| ligation name | 728 + 881 |
| volume of vector | 2,68 |
| volume of insert | 5,32 |
| T4 ligase buffer (10x) | 1 |
| T4 ligase | 1 |
Transformation:
Transformation was performed according to standard protocol using BL21 cells.
RT-PCR
Investigator: Kira
For further experiments, RNA has to be translated into cDNA. The PCR was performed according to the manufacturer protocol.
153. labday 18.10.2010
quantitative real-time PCR for detection of virus titer
Investigator: Achim
- qPCR of harvested virus particles to determine the virus titers of our different constructs
- Total number of samples: 58
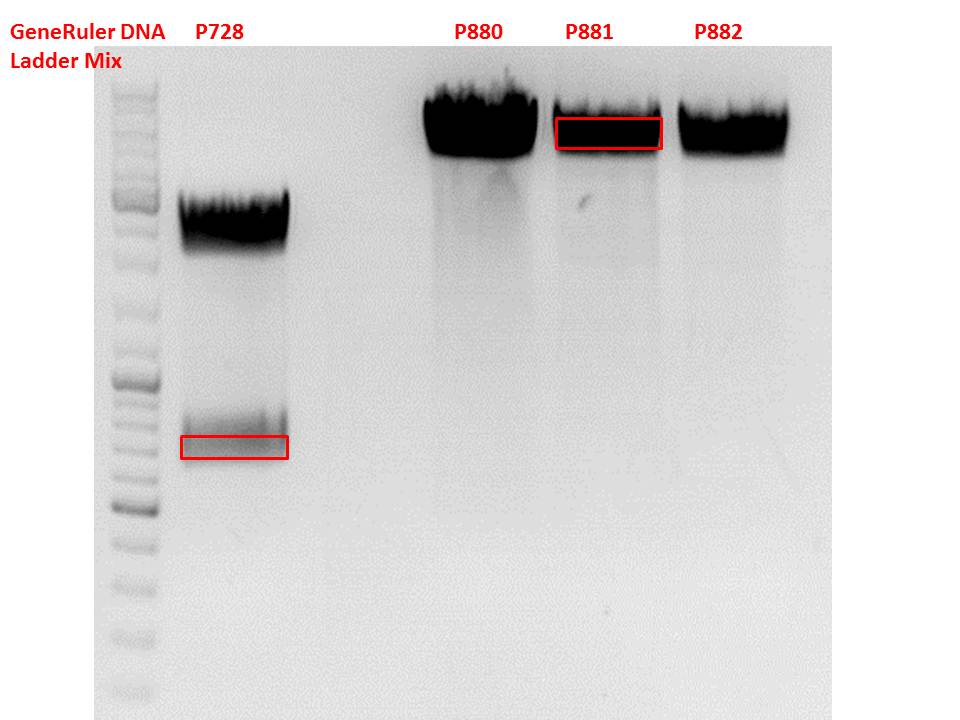
Test-digestion of pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_hGH_rITR_SDM-PstI clone 1, 2 and 3 (P877, 878 and 879)
Investigator Patrick
Check the plasmid for leftITR:
0,5 µl EcoRI, 1 µl BstEII, 7 µl DNA, 2 µl Buffer 4, 2 µl BSA, 7,5 µl H2O, 45 minutes 37°C, 45 minutes 60°C
Check the plasmid for hGH_rITR and ... :
0,5 µl AgeI, 0,5 µl PstI, 3 µl DNA, 1 µl BSA, 1 µl Buffer 4, 4 µl H2O, 70 minutes 37°C
Expected results:
- leftITR: about 190 bp
- hGH_rITR: about 670 bp
Unfortunately the digestions had to be reapeated because i didnt switch on the current so the samples and especially the 1kb GeneRuler marker diffused.
The second run: see above
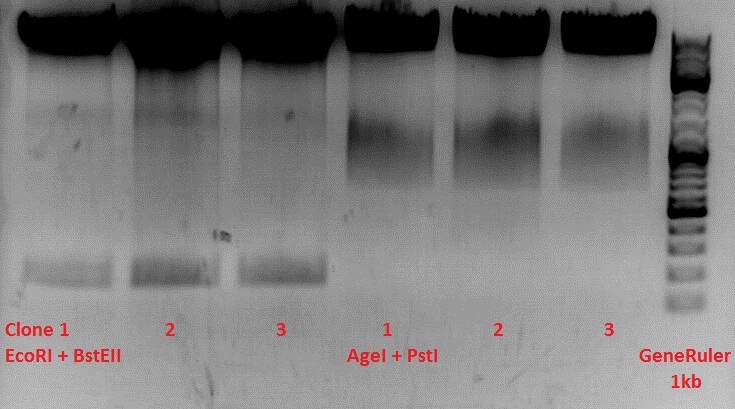
PstI or BstEII seems to work not properly
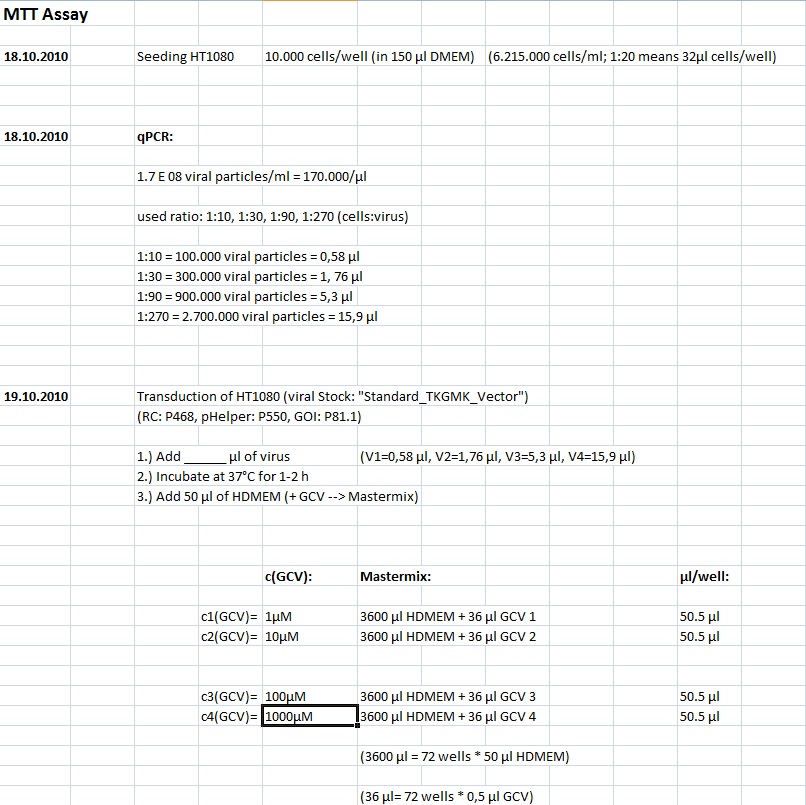
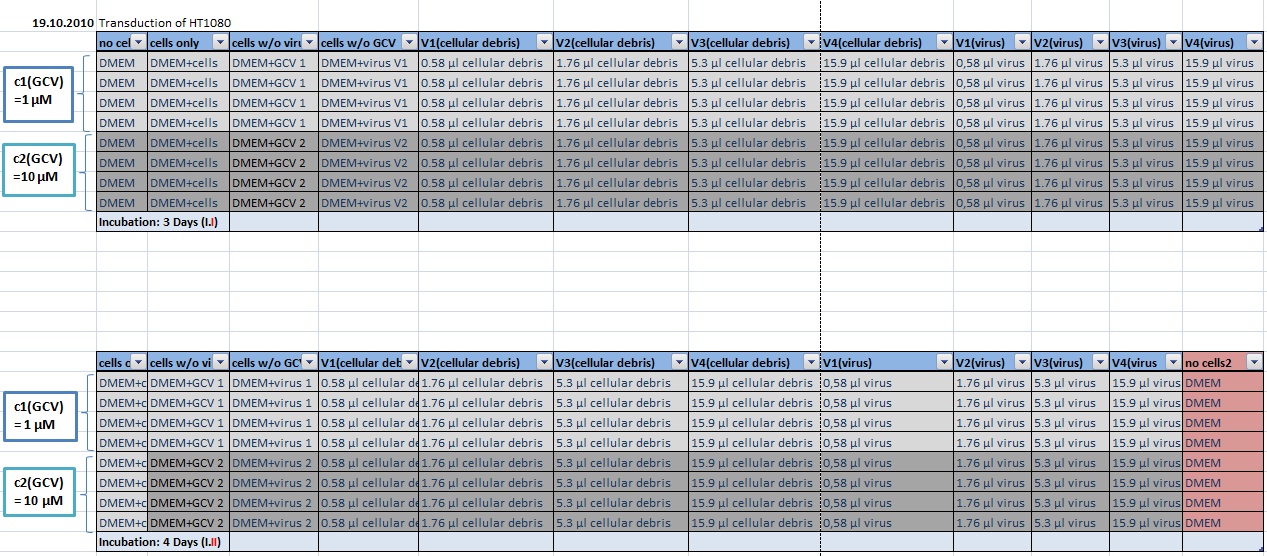
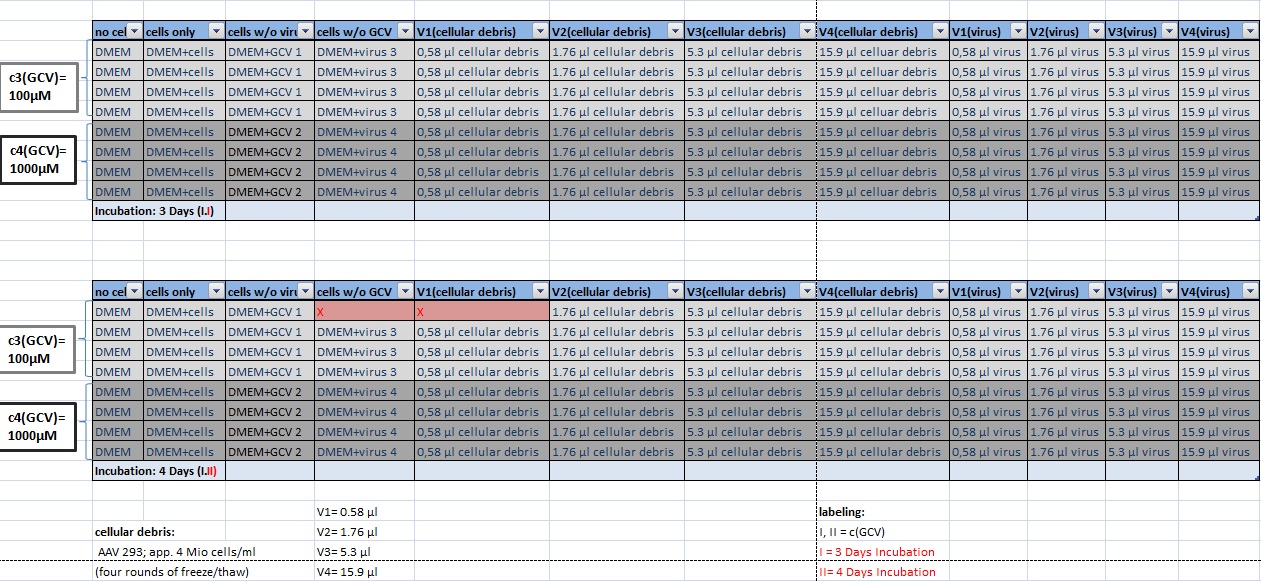
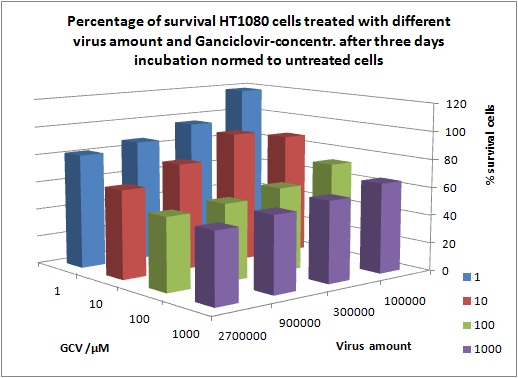
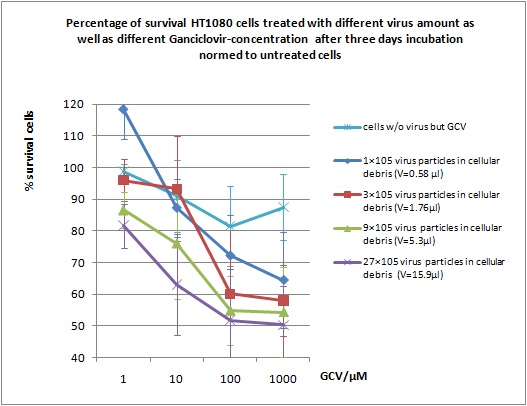
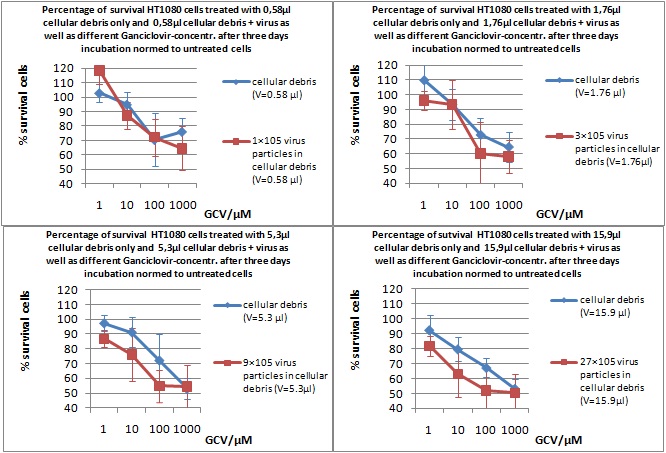
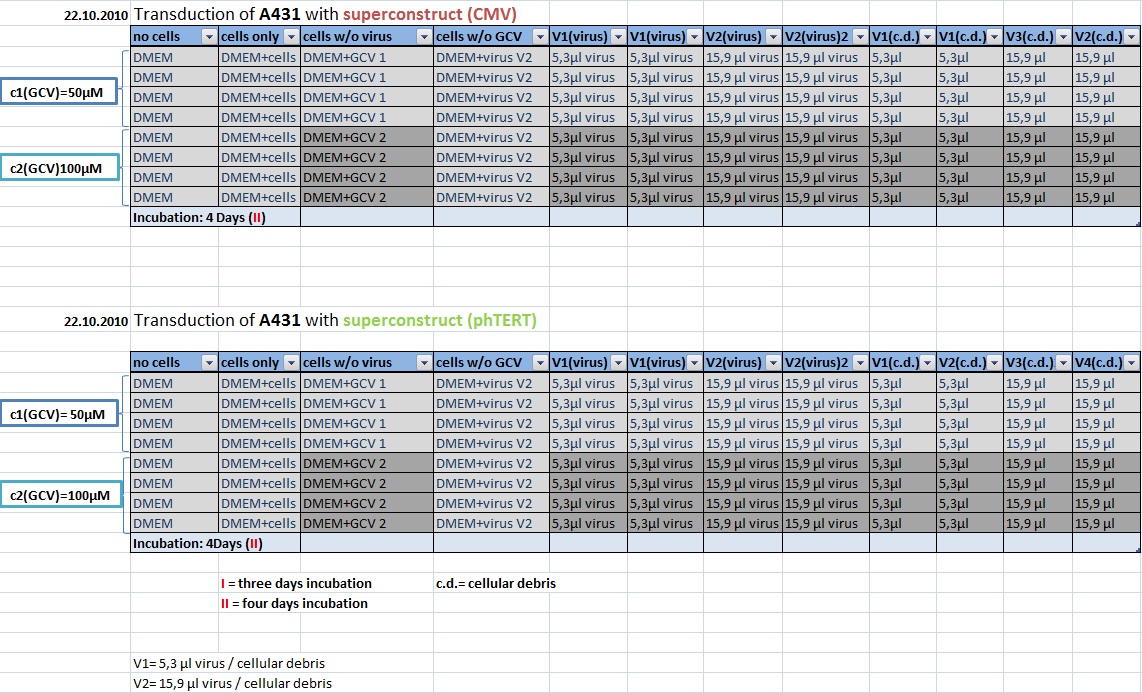
MTT Assay
Investigator Kerstin, Anissa
Results:
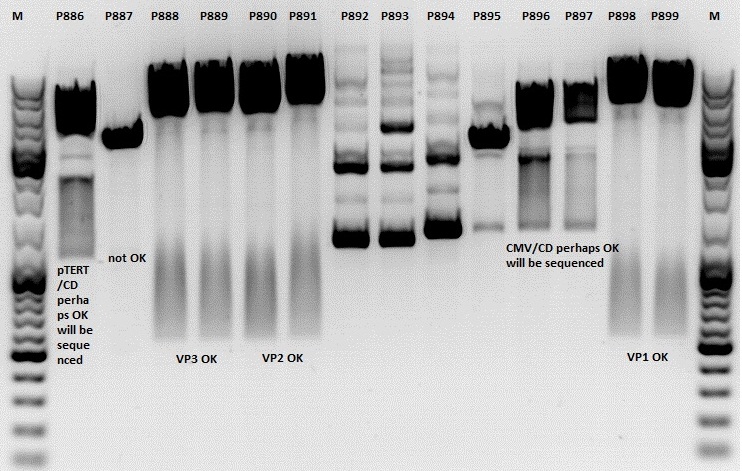
Mini-Prep and test digestion of several constructs
Investigator: Jessica
Glycerol stocks were prepared:
- B702 = pSB1C3_lITR_pTERT_ßglobin_CD_hGH_rITR clone 1
- B703 = pSB1C3_lITR_pTERT_ßglobin_CD_hGH_rITR clone 2
- B704 = pSB1C3_001_VP3 clone 1
- B705 = pSB1C3_001_VP3 clone 2
- B706 = pSB1C3_001_VP2 clone 1
- B707 = pSB1C3_001_VP2 clone 2
- B708 = pSB1C3_001_Rep78 clone 1
- B709 = pSB1C3_001_Rep78 clone 2
- B710 = pSB1C3_001_Rep52 clone 1
- B711 = pSB1C3_001_Rep52 clone 2
- B712 = pSB1C3_lITR_CMV_ßglobin_CD_hGH_rITR clone 1
- B713 = pSB1C3_lITR_CMV_ßglobin_CD_hGH_rITR clone 2
- B714 = pSB1C3_001_VP1 clone 1
- B715 = pSB1C3_001_VP1 clone 2
Mini-Prep was performed according to standard protocol:
- P886 = pSB1C3_lITR_pTERT_ßglobin_CD_hGH_rITR clone 1 c= 232,2ng/µl
- P887 = pSB1C3_lITR_pTERT_ßglobin_CD_hGH_rITR clone 2 c= 186,2ng/µl
- P888 = pSB1C3_001_VP3 clone 1 c= 300,8ng/µl
- P889 = pSB1C3_001_VP3 clone 2 c= 284,4ng/µl
- P890 = pSB1C3_001_VP2 clone 1 c= 298,3ng/µl
- P891 = pSB1C3_001_VP2 clone 2 c= 299,9ng/µl
- P892 = pSB1C3_001_Rep78 clone 1 c= 143,9ng/µl
- P893 = pSB1C3_001_Rep78 clone 2 c= 163,4ng/µl
- P894 = pSB1C3_001_Rep52 clone 1 c= 166,6ng/µl
- P895 = pSB1C3_001_Rep52 clone 2 c= 181,6ng/µl
- P896 = pSB1C3_lITR_CMV_ßglobin_CD_hGH_rITR clone 1 c= 250,5ng/µl
- P897 = pSB1C3_lITR_CMV_ßglobin_CD_hGH_rITR clone 2 c= 173,4ng/µl
- P898 = pSB1C3_001_VP1 clone 1 c= 272,7ng/µl
- P899 = pSB1C3_001_VP1 clone 2 c= 294,8ng/µl
- P900 = pSB1C3_hGH_rITR (from B160) c= 136,7ng/µl
Test digestion:
| Components | P886,887,892,893,894,895,896,897 / µl | P888,889,890,891898,899 / µl |
| DNA | 1,5 | 1,5 |
| Buffer | (4) 1 | (2) 1 |
| BSA (10x) | 1 | 1 |
| NgoMIV | 0,4 | - |
| XbaI | 0,4 | - |
| PstI | - | 0,6 |
| XcmI | - | 0,4 |
| H2O | 4,5 | 4,5 |
| Total volume | 10 | 10 |
Gel:
1,0g agarose, 100 ml TAE (1%), 6 µl GELRED, Volt, running time minutes
Comment: Rep 52/78 will be checked,pSB1C3_lITR_CMV_ßglobin_CD_hGH_rITR and pSB1C3_lITR_pTERT_ßglobin_CD_hGH_rITR will be sequenced
PCR of mGMK and SR39
Investigator: Anna
Plasmids:
pSB1C3_mGMK_TK30_SDM-PstI clone 2(P804)
pSB1C3_mGMK_sr39 clone 1(P860)
Oligos:
O193: pTK30_for
O81: pmgmk_tk30_suffix_RFC25_rev
PCR Mix:
| Components | Volume /µl |
| Phusion Buffer | 10 |
| dNTP | 1 |
| Primer_for | 2,5 |
| Primer_rev | 2,5 |
| DNA template | 1 |
| H2O | 32,5 |
| Total volume | 50 |
PCR Program:
| Cycles | Temperature | Time |
| 98°C | 60 sec | |
| 98°C | 15 sec | |
| 8x | 52°C | 25 sec |
| 72°C | 25 sec | |
| 98°C | 15 sec | |
| 17x | 67°C | 25 sec |
| 72°C | 25 sec | |
| 1x | 72°C | 5 min |
| Hold 4°C |
Gel:
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 115 Volt
[[Image:|550px|]]
154. labday 19.10.2010
Midi-Prep
Investigator: Chris W.
Midi-Prep of:
pSB1C3_lITR_CMV_betaglobin_mVenus_hGH_rITR clone1 =P901 =B200
pSB1C3_lITR_phTERT_betaglobin_mGMK_TK30_hGH_rITR_SDM-PstI clone 2 =P902 =B697
The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| plasmid-no. | P901 | P902 |
| concentration (ng/µl) | 1563,63 | 1348,26 |
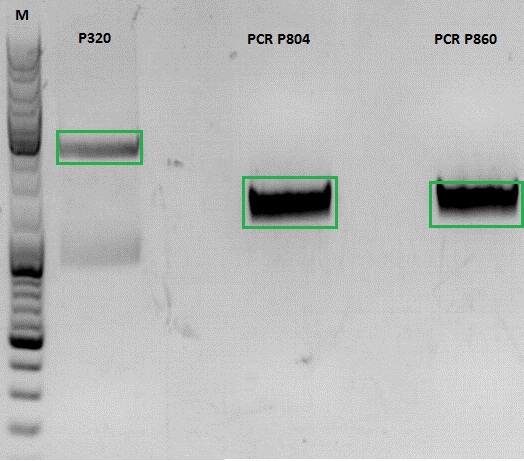
Continuation of PCR of mGMK and SR39
Investigator: Jessica
- vector (P320) and PCR product was digested
| Components | P320 / µl | PCR product P804 and P860 / µl |
| DNA | 1,5 | 20 |
| Buffer | 2 | 3 |
| BSA (10x) | 2 | 3 |
| AgeI | 1 | 1,5 |
| XbaI | 1 | 1,5 |
| H2O | 14,5 | 1 |
| Total volume | 20 | 30 |
Ligation
- P320 c= 5,08 ng/µl
- P804 c= 11,73 ng/µl
- P860 c= 26,57 ng/µl
- P320 + P804: 4,93µl : 3,07µl
- P320 + P860: 6,28µl : 1,72µl
Transformation with BL21 and Cm
Cloning of CMV into pSB1C3_001_VP1, pSB1C3_001_VP2 and pSB1C3_001_VP3
Investigator: Kerstin, Anna
Plasmids:
- P888: pSB1C3_001_VP3, c = 300,8 ng/µl
- P890: pSB1C3_001_VP2, c = 298,3 ng/µl
- P898: pSB1C3_001_VP1, c = 272,7 ng/µl
- P727: pSB1C3_001_CMV, c = 225,5 ng/µl
Digestion:
| components | VP3 | VP2 | VP1 | CMV |
| DNA | 4 | 4 | 4 | 8 |
| BSA (10x) | 2 | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 | 2 |
| Enzyme EcoI | 1 | 1 | 1 | 1 |
| Enzyme XbaI | 1 | 1 | 1 | - |
| Enzyme SpeI | - | - | - | 1 |
| H2O | 10 | 10 | 10 | 10 |
| Total | 20 | 20 | 20 |
- Digestion: 2h @ 37°C
Gel:
- 1% agarose gel, 1 µl Gelred, run for 45 min
Gel extraction
| sample name | VP3 | VP2 | VP1 | CMV |
| nanodrop concentrations | 44,36 | 32,33 | 22,73 | 18,5 |
| expected fragment size | 4100 | 4000 | 3700 | 650 |
Ligation:
| ligation name | VP3 + CMV | VP2 + CMV | VP1 + CMV |
| volume of vector | 5,7 | 4,3 | 5 |
| volume of insert | 3,3 | 3,7 | 3 |
| T4 ligase buffer (10x) | 1 | 1 | 1 |
| T4 ligase | 1 | 1 | 1 |
- Ligation @ RT for 30 min
Trafo:
Was done following the standard protocol using BL21 cells.
155. labday 20.10.2010
Midi-Prep
Investigator: Chris W.
Midi-Prep of:
pSB1C3_lITR_pTERT_ßglobin_CD_hGH_rITR clone 1 =P903 =B702
pSB1C3_lITR_CMV_ßglobin_CD_hGH_rITR clone 1 =P904 =B712
The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| plasmid-no. | P903 | P904 |
| concentration (ng/µl) | 832,63 | 1174,49 |
156. labday 21.10.2010
ÄKTA Chromatography and Ultrafiltration of virus particles
Investigator: Hanna
ÄKTA chromatography with VP1up_NLS_mVenus_VP2/3 containing virus particles was conducted. Fraction 5 - 10 delivered highest protein concentrations.
| Sample | A(260 nm) | A(280 nm) | A(515 nm) (YFP) |
| 5 | 0.032 | 0.027 | 0.003 |
| 6 | 0.019 | 0.019 | 0.003 |
| 7 | 0.075 | 0.09 | 0.01 |
| 8 | 0.054 | 0.075 | 0.007 |
| 10 | -0.008 | -0.008 | 0.005 |
A further attempt was conducted which included digestion with Benzonase (1 hour) prior to ÄKTA chromatography. Following protein concentrations were obtained:
| Sample | A(260 nm) | A(280 nm) | A(515 nm) (YFP) |
| 5 | 0.005 | 0.007 | 0.003 |
| 6 | 0.047 | 0.041 | 0.006 |
| 7 | 0.151 | 0.153 | 0.01 |
| 8 | 0.172 | 0.2 | 0.009 |
| 9 | 0.098 | 0.128 | 0.009 |
| 10 | 0.053 | 0.074 | 0.005 |
Ultrafiltration
Ultrafiltration of CFP_MiddleLinker_VP2/3 containing virus particles and 587-BAP virus particles were concentrated via Vivaspin-Ultrafiltration:
- 20 mL virus containing cell culture supernatant was added to GE Vivaspin 20 filter and centrifuged with 4000 g at 15°C until 750 - 1000 µL was left-
- 5 mL Bis-Trus buffer (pH 6) was added and centrifuged again with 4000 g at 15°C (washing).
- This step was repeated 3 more times.
- Membrane was carefully resuspended and cleared. Suspension was transfered to low-binding eppi and centrifuged with 10000 g for 10 minutes at 15°C.
- Supernatant was transfered to new low-binding eppi and again centrifuged with 10000 g for 10 minutes at 15°C.
- Supernatant was transfered to new low-binding eppi and stored at 4°C over night. To do: ÄKTA chromatography.
MTT Assay: Testing Superconstructs
Investigator: Anissa, Kerstin
157. labday 22.10.2010
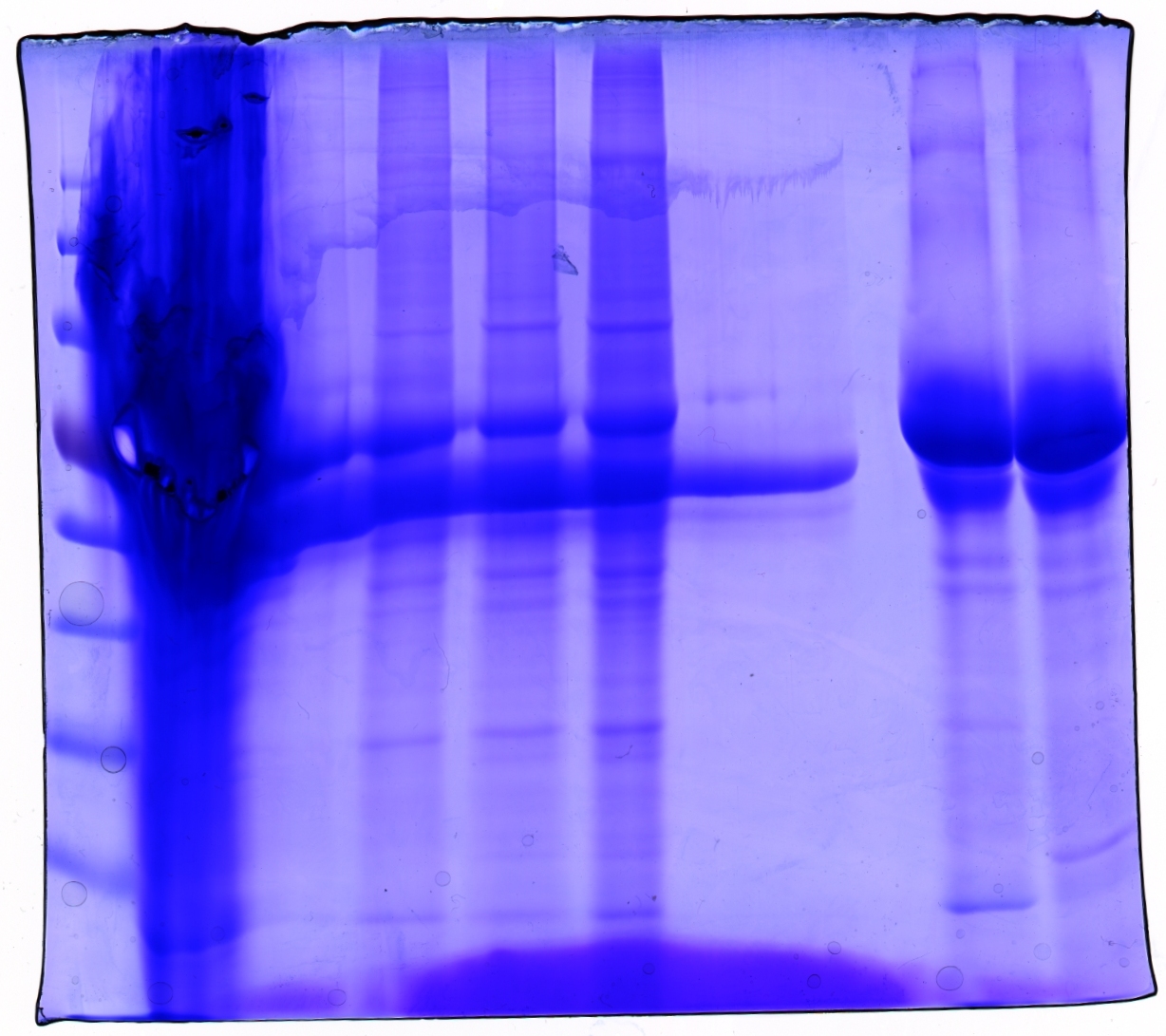
SDS PAGE and Coomassie staining
Investigator: Hanna
Prior to performing Western Blot we decided to investigate running behaviour of different samples.
- 1. Cell debris (control)
- 2. Cell debris containing virus particles
- 3. Concentrated virus stock (containing CFP_MiddleLinker_VP2/3)
- 4. ÄKTA purified virus stock: Fraction 6
- 5. ÄKTA purified virus stock: Fraction 7
- 6. Benzonase treated, ÄKTA purified virus stock: Fraction 7
- 7. Benzonase treated, ÄKTA purified virus stock: Fraction 8
5 µL Laemmli buffer was added to 20 µL sample. Samples were incubated at 95°C for 8 minutes and loaded onto a SDS gel (10 %). SDS PAGE was performed at 90 V (collection gel) resp. 120 V (separation gel).
Gel was put into Coomassie dye, heated for 30 seconds in microwave and incubated for 1 hours shaking.
Gel was decolorized in acetic acid (20%).
Loading plan:
| Marker | Concentrated Stock | ÄKTA 6 | ÄKTA 7 | Benzonase/ÄKTA 7 | Benzonase/ÄKTA 8 | - - - | - - - | Cell debris | Cell debris + Virus |
Gel picture shows that concentration works :)
In addition to that one can see that the BSA bands disappear after ÄKTA chromatography.
Next step: Western Blot of ÄKTA purified virus stocks.
158. labday 23.10.2010
159. labday 24.10.2010
Midi-Prep
Investigator: Chris W.
Midi-Prep of:
pSB1C3_001_RC_IRCK_VP2-ko_HSPG-ko_P5tataless cl1 =P966 =B523
The Midi-Prep were performed according to the standard protocol yielding the following concentration:
| plasmid-no. | P966 |
| concentration (ng/µl) | 310,69 |
160. labday 25.10.2010
161. labday 26.10.2010
162. labday 27.10.2010
163. labday 28.10.2010
164. labday 29.10.2010
165. labday 30.10.2010
166. labday 31.10.2010
 "
"