Team:Newcastle/26 August 2010
From 2010.igem.org

| |||||||||||||
| |||||||||||||
Contents |
yneA
PCR (Repeat)
Aim
To repeat the PCR that we did yesterday using the correct rocF primers.
Materials and Protocol
Please refer to PCR.
PCR Purification
Aim
To remove unwanted primers, taq polymerase, buffer and salts to obtain pure DNA.
Materials and Protocol
Please refer to PCR purification.
Restriction Digest
Aim
To digest the PCR products of pSB1C3 and yneA from PCR purification with enzymes EcoR1 and Nhe1.
Materials and Protocol
Please refer to restriction digest.
Results, Discussion and Conclusion
We run the digested products with gel electrophoresis to determine whether the digest worked.
Gel extraction
Aim
To purify the DNA of yneA and pSB1C3 by extracting the bands from the gel after running gel electrophoresis. Concentration of DNA is then checked with NanoDrop.
Materials and Protocol
Please refer to:
Results
The gel did not show any band when we looked at it from GelDoc.
Conclusion
The restriction digest did not work, so we will repeat the protocol again tomorrow.
First transformation of 'Bacillius subtilis 168' with Prrnb-GFP containing YneA
Aim
The aim of the experiment is to perform insert the plasmid Prrnb-GFP containing YneA which have been ligated eariler into the chromosome of 'Bacillus subtilis 168'. 'B. subtilis' containing the intergated vector will be resistance to both the antibiotic chloramphenicol and streptomycin, therefore those that have successful intergated will be selected with agar plates that contain these antibiotic. The second step will be to identify those colones that have the plasmid intergated at the corerct position in the chromosome, which is the amylase locus. Thus those that have intergardted at the wrong position will not be able to break down starch, which can be tested with the iodine test.
Materials and Protocol
Please refer to: Transformation of Bacillus subtilis Note: Overnigth culture of 'B. subtilis 168' in MM competence medium was done the day before and the iodine test was performed the day after.
Results
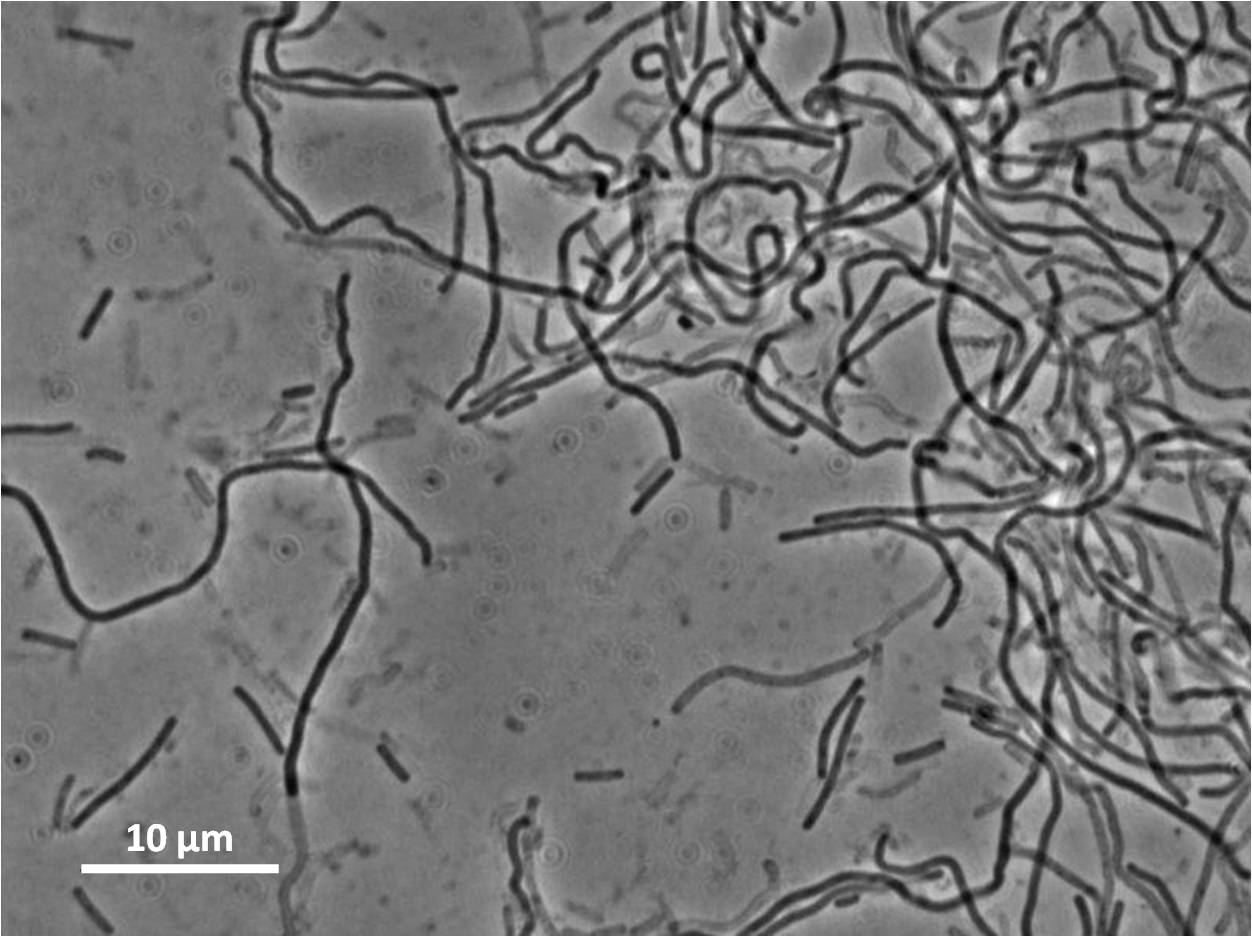
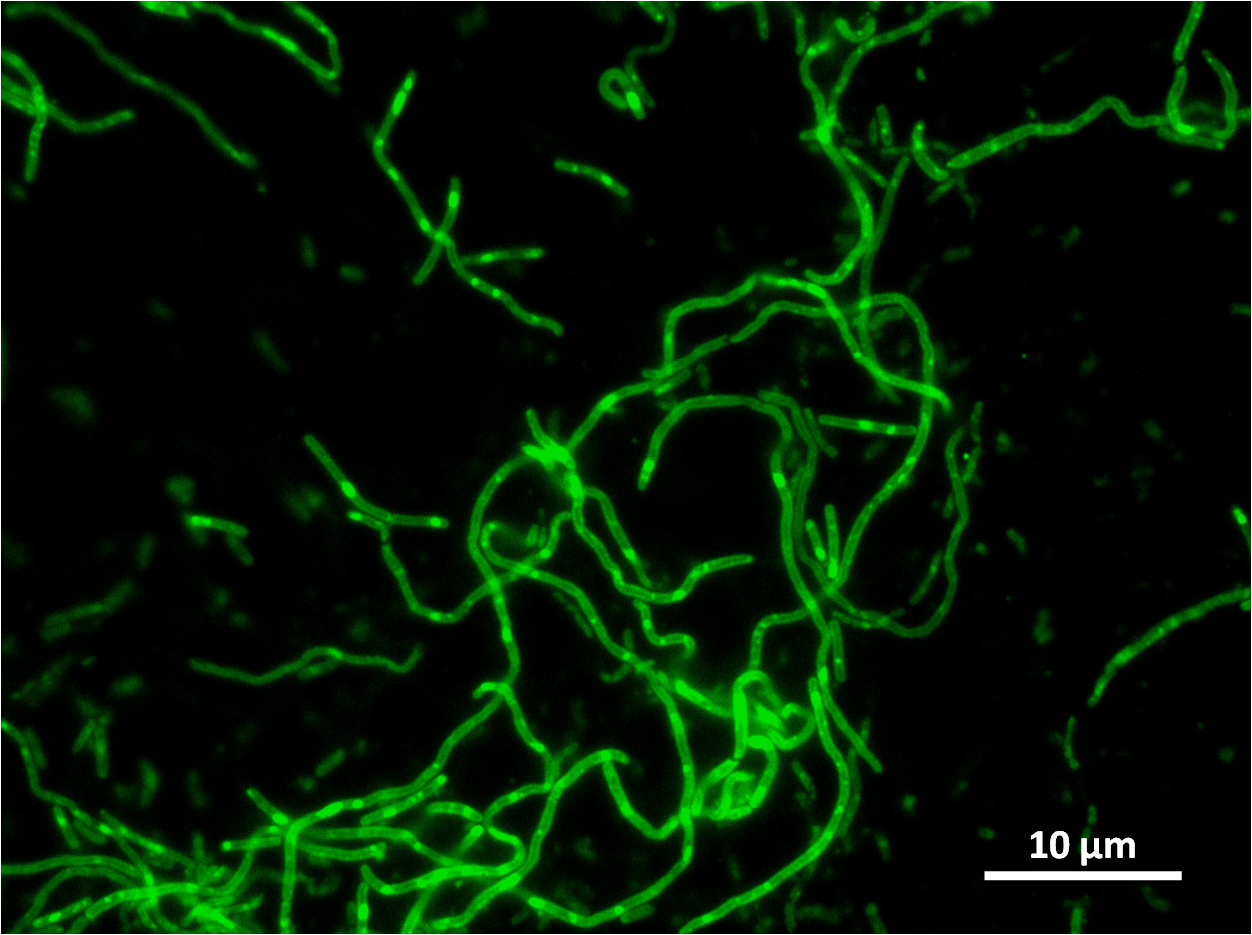
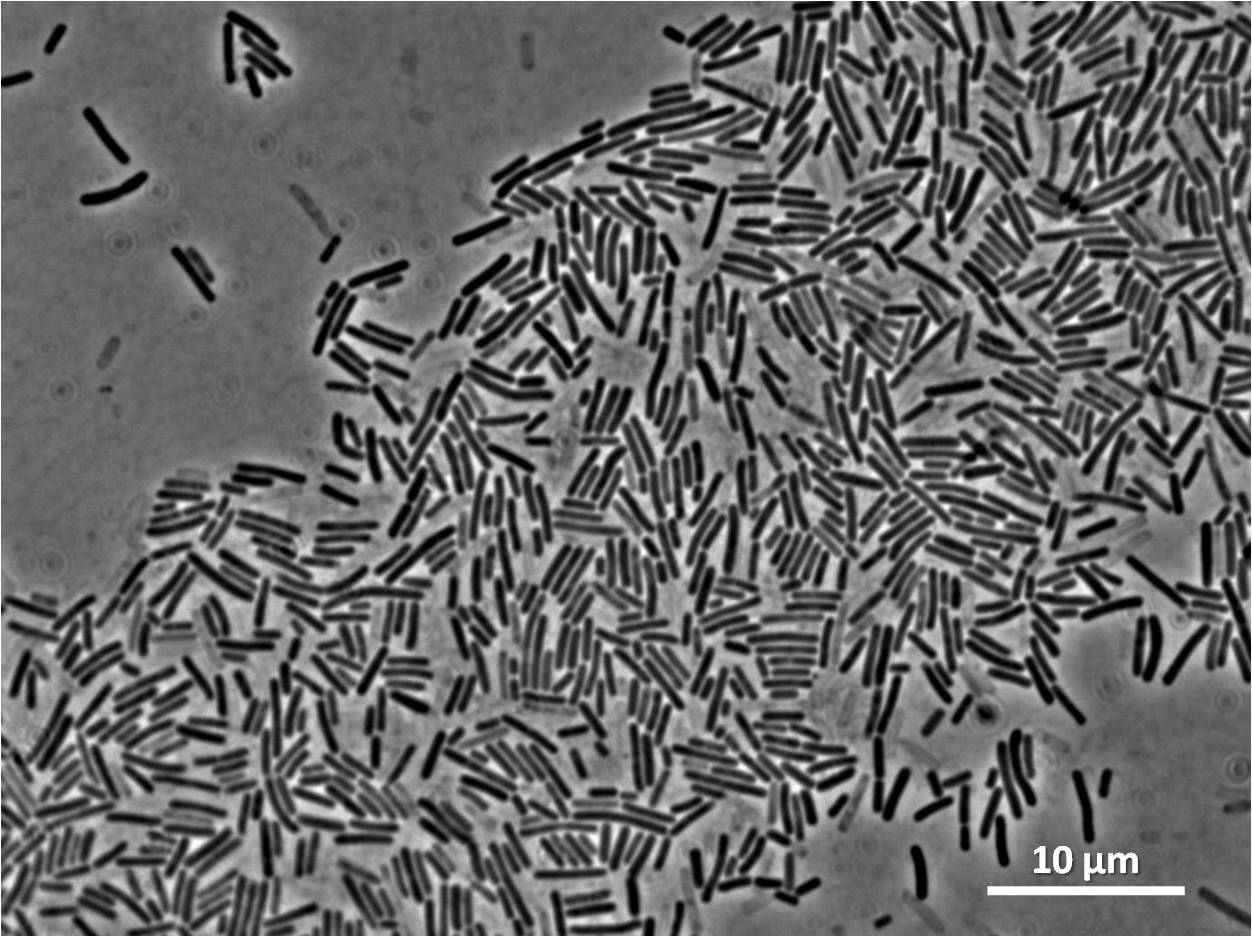
Unfortunately our colonies had halos. Although the insert has integrated, the colonies have antibiotic restistance, the insert has not integrated at the amyE locus.
However when we checked the colonies under a microscope the cells were filamentous due to integration elsewhere on the Bacillus subtilis 168 chromosome perhaps at the native yneA locus.
 
|
 "
"