Team:Heidelberg/Project/miRNA Kit
From 2010.igem.org
(→miTuner Kit components) |
(→miTuner Kit components) |
||
| Line 33: | Line 33: | ||
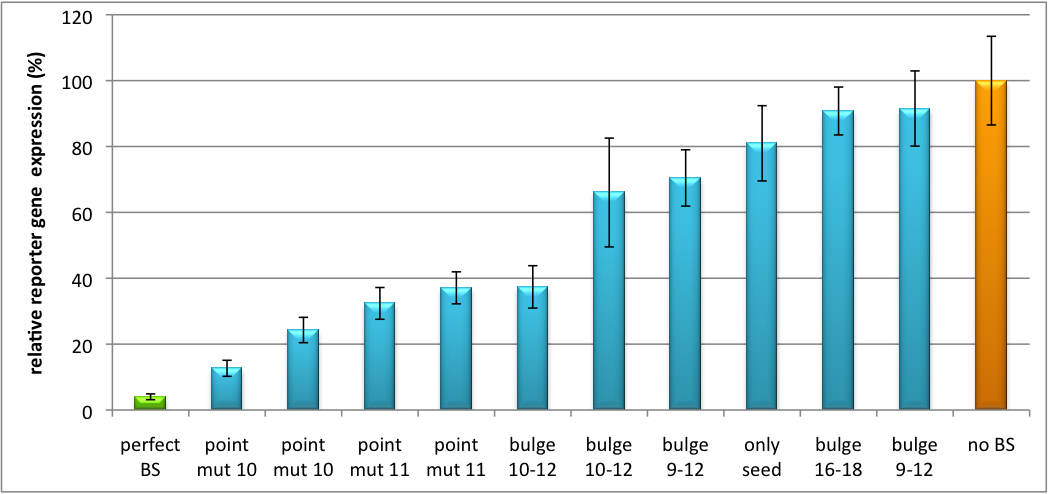
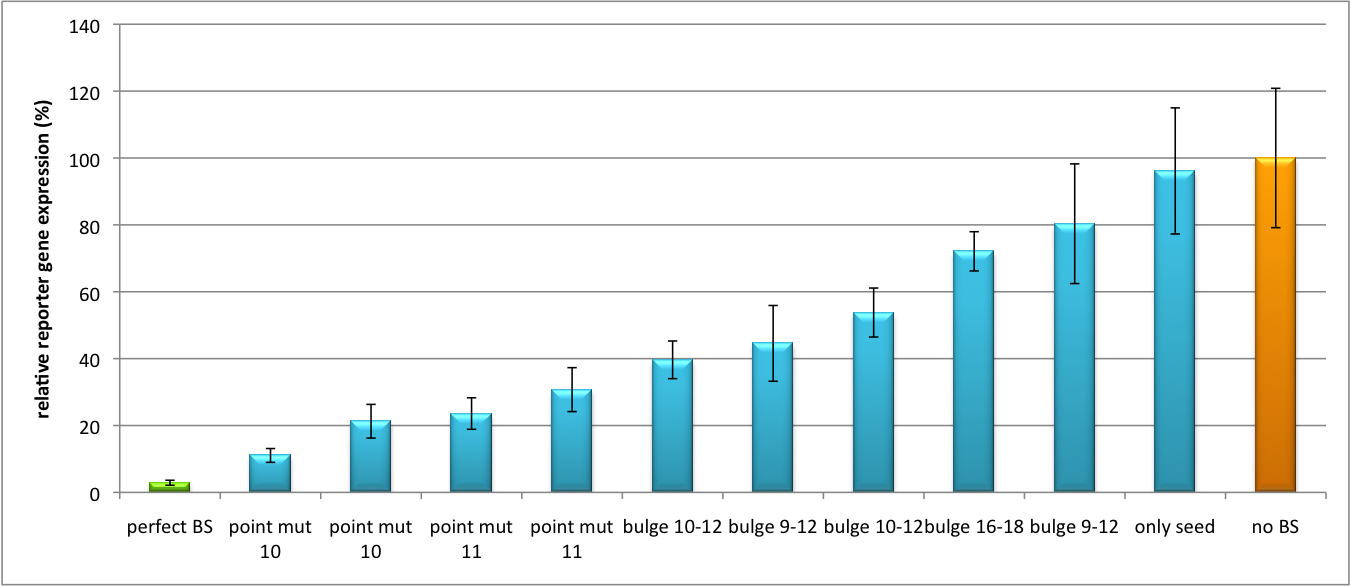
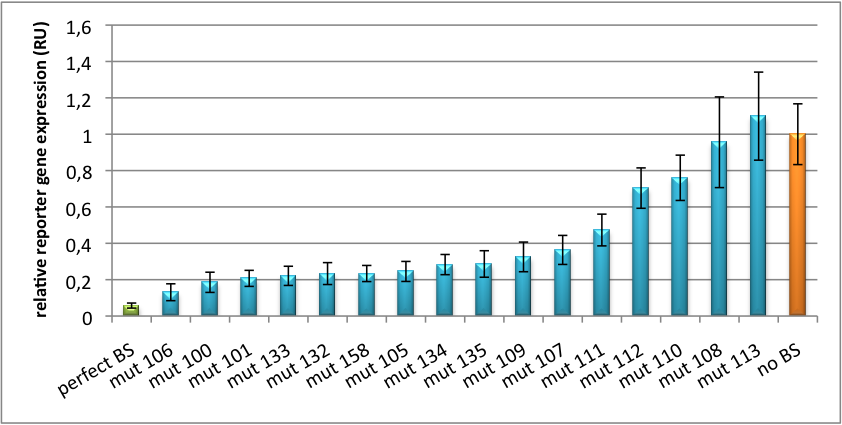
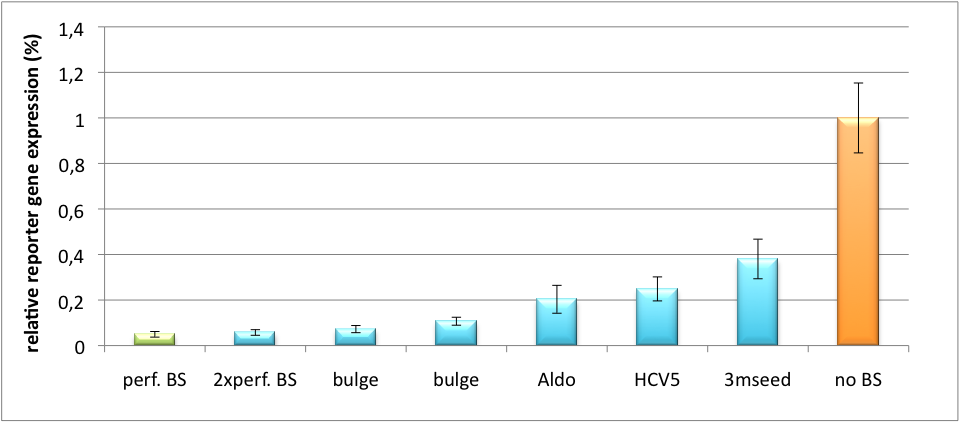
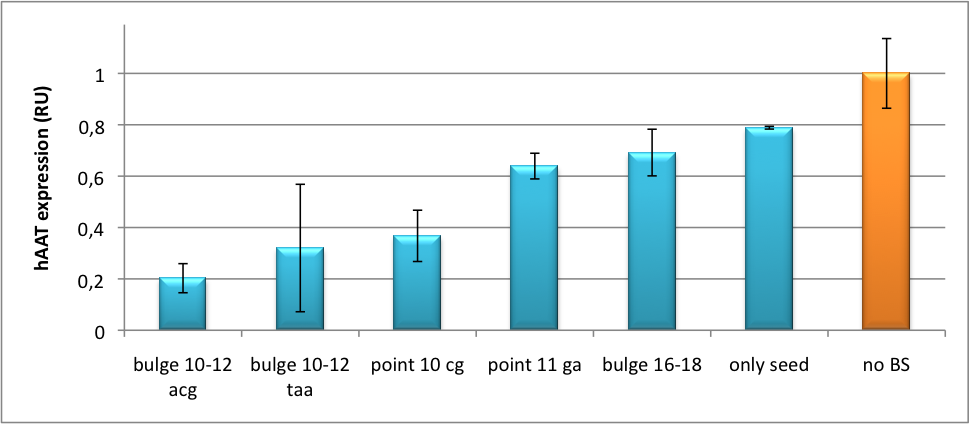
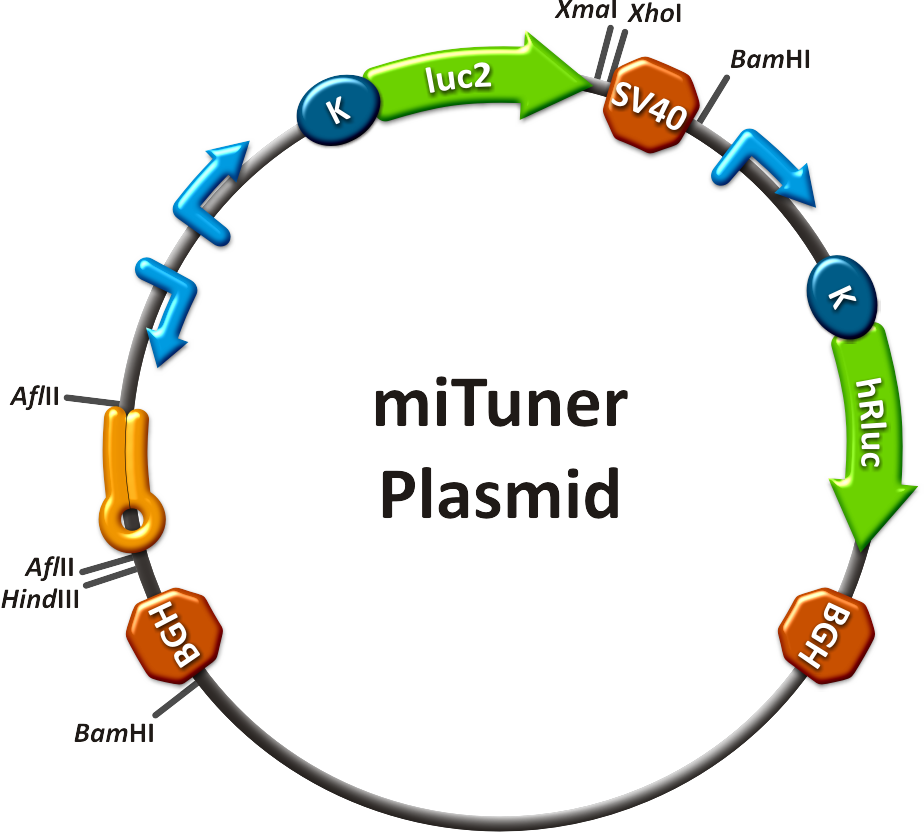
Our '''synthetic miRNA Kit''' guarantees at least for individually modifiable but still ready-to-use constructs to interfere genetic circuits with synthetic or endogenous miRNAs. We preciously show, that gene expression can thereby by adjusted - tuned - to an arbitrary level. The '''miTuner''' (see sidebar) allows on the simultaneous expression of a synthetic miRNA and a gene of interest that is fused with a designed binding site for this specific miRNA. Our modular kit comes with different parts that can be combined by choice, e. g. different mammalian promoters and characterized binding sites of specific properties. By choosing a certain binding site to tag the GOI, one can tune the expression of this gene. Depending on the GOI, different means for read out of gene expression come into play. At first, we applied [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Dual_Luciferase_Assay dual-luciferase assay], since we used Luciferase as a reporter for a proof-of-principle approach. Later on, semi-quantitative immunoblots were prepared for testing of therapeutic genes. However, all the received information fed our models, thereby creating an '''integrative feedback loop between experiments and in silico simulation'''. | Our '''synthetic miRNA Kit''' guarantees at least for individually modifiable but still ready-to-use constructs to interfere genetic circuits with synthetic or endogenous miRNAs. We preciously show, that gene expression can thereby by adjusted - tuned - to an arbitrary level. The '''miTuner''' (see sidebar) allows on the simultaneous expression of a synthetic miRNA and a gene of interest that is fused with a designed binding site for this specific miRNA. Our modular kit comes with different parts that can be combined by choice, e. g. different mammalian promoters and characterized binding sites of specific properties. By choosing a certain binding site to tag the GOI, one can tune the expression of this gene. Depending on the GOI, different means for read out of gene expression come into play. At first, we applied [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Dual_Luciferase_Assay dual-luciferase assay], since we used Luciferase as a reporter for a proof-of-principle approach. Later on, semi-quantitative immunoblots were prepared for testing of therapeutic genes. However, all the received information fed our models, thereby creating an '''integrative feedback loop between experiments and in silico simulation'''. | ||
| - | == miTuner Kit components == | + | == miTuner Kit components == <br /> |
| - | The miTuner Kit consists of three basic components: | + | The miTuner Kit consists of three basic components: <br /> |
| - | a)A kit of standardized synthetic microRNAs, corresponding binding sites, promoters and luciferase expression constructs as well as measurement constructs assembled in the BB-2 standard. As the miTuner kit was enabled | + | a)A kit of standardized synthetic microRNAs, corresponding binding sites, promoters and luciferase expression constructs as well as measurement constructs assembled in the BB-2 standard. As the miTuner kit was enabled <br /> |
b)Protocols for engineering synthetic microRNAs, synthetic single microRNA binding sites as well as microRNA binding site patterns | b)Protocols for engineering synthetic microRNAs, synthetic single microRNA binding sites as well as microRNA binding site patterns | ||
| - | + | <br /> | |
Please find further information about the kit componenets and engineering of the kit [[2010.igem.org/wiki/index.php?title=Team:Heidelberg/Project/miRNA_Kit|here]]. | Please find further information about the kit componenets and engineering of the kit [[2010.igem.org/wiki/index.php?title=Team:Heidelberg/Project/miRNA_Kit|here]]. | ||
Revision as of 20:11, 27 October 2010

|
|
||
 "
"