Team:Heidelberg/Project/miRNA Kit
From 2010.igem.org
(→In Vitro Regulation of a Therapeutic Gene, HAAT) |
(→In Vitro Regulation of a Therapeutic Gene, hAAT) |
||
| Line 92: | Line 92: | ||
=== <i>In Vitro</i> Regulation of a Therapeutic Gene, hAAT === | === <i>In Vitro</i> Regulation of a Therapeutic Gene, hAAT === | ||
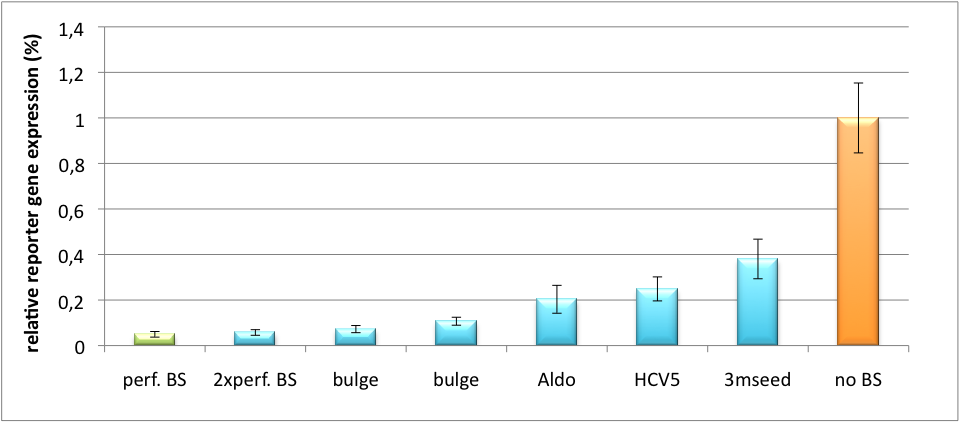
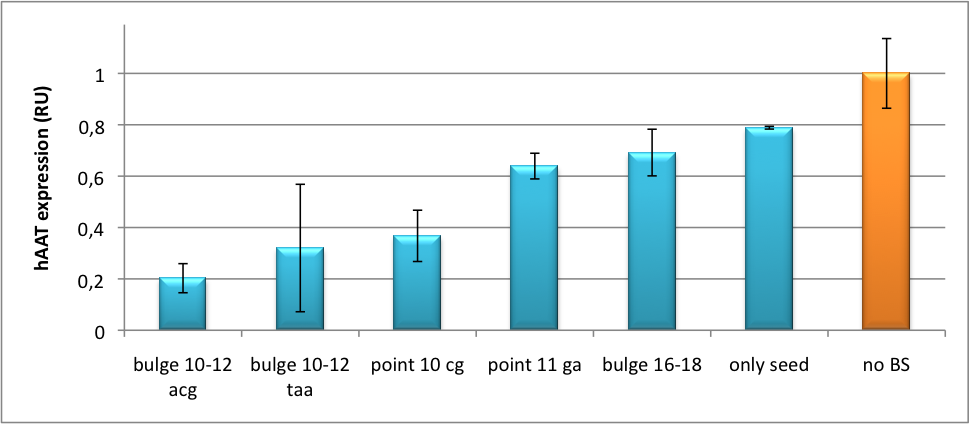
| - | We further tested our kit using a gene that is an interesting candidate for gene therapy, i. e. human alpha-1-antitrypsin (hAAT). Tight control of the genetic activity is fundamental, since deficiencies of hAAT can cause emphysema {{HDref| Lu et al., 2006}}. With our tuning kit we have a powerful mean at hand to mediate expression levels. In this approach, we tagged hAAT, that we used as our GOI, with binding sites (for miRsAg) that we measured and characterized with our [https://2010.igem.org/Team:Heidelberg/Project/miMeasure miMeasure] construct beforehand (data not shown). There is some evidence, that the principle works also with this therapeutic gene in HeLa cells (fig. 6). This is a first potential therapeutic approach applying [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#ELISA ELISA] for measurements. | + | We further tested our kit using a gene that is an interesting candidate for gene therapy, i. e. human alpha-1-antitrypsin (hAAT). Tight control of the genetic activity is fundamental, since deficiencies of hAAT can cause emphysema {{HDref|Lu et al., 2006}}. With our tuning kit we have a powerful mean at hand to mediate expression levels. In this approach, we tagged hAAT, that we used as our GOI, with binding sites (for miRsAg) that we measured and characterized with our [https://2010.igem.org/Team:Heidelberg/Project/miMeasure miMeasure] construct beforehand (data not shown). There is some evidence, that the principle works also with this therapeutic gene in HeLa cells (fig. 6). This is a first potential therapeutic approach applying [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#ELISA ELISA] for measurements. |
[[Image:HAAT ELISA.png|thumb|center|400px|'''Figure 6: hAAT expression in relative units depending of binding site properties.''' SV40 driven hAAT was fused to binding sites for miRsAg that was expressed from a co-transfected plasmid in HeLa cells.]] | [[Image:HAAT ELISA.png|thumb|center|400px|'''Figure 6: hAAT expression in relative units depending of binding site properties.''' SV40 driven hAAT was fused to binding sites for miRsAg that was expressed from a co-transfected plasmid in HeLa cells.]] | ||
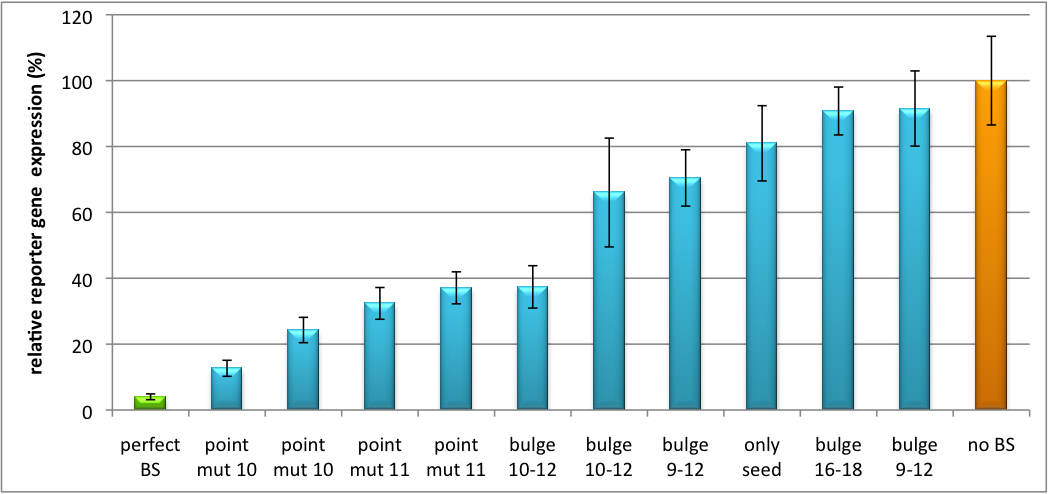
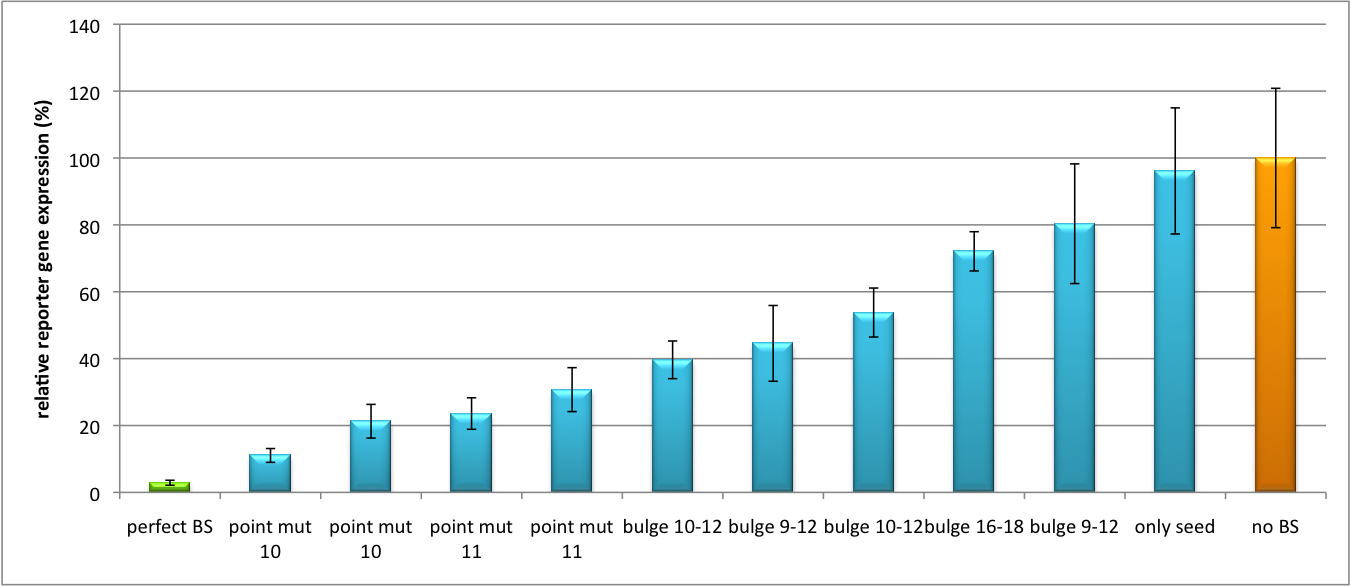
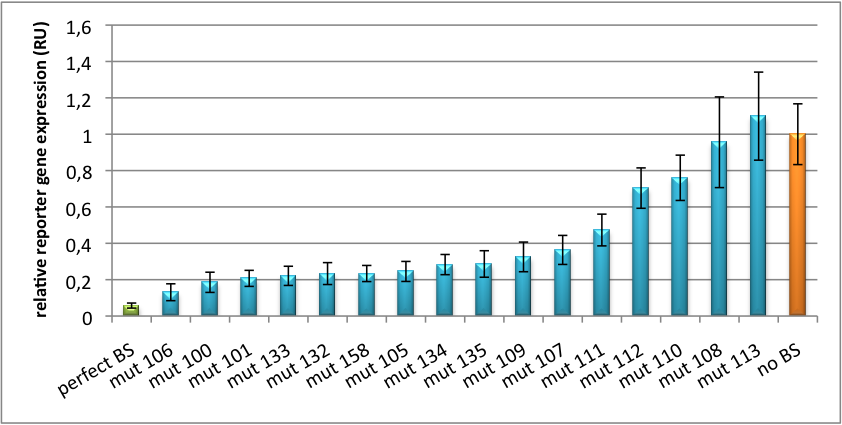
It is obvious: different binding sites result in different knockdowns of gene expression. Some imperfect binding sites - e. g. single seed region - indicate even similar expression levels in accordance to the figures shown before. It can be stated, that the tuning idea seems to work for attempts varying in applied miRNAs, binding sites and reporter genes. | It is obvious: different binding sites result in different knockdowns of gene expression. Some imperfect binding sites - e. g. single seed region - indicate even similar expression levels in accordance to the figures shown before. It can be stated, that the tuning idea seems to work for attempts varying in applied miRNAs, binding sites and reporter genes. | ||
Revision as of 20:04, 27 October 2010

|
|
||
 "
"