Team:Heidelberg/Project/miRNA Kit
From 2010.igem.org
Laura Nadine (Talk | contribs) (→Results) |
Laura Nadine (Talk | contribs) |
||
| Line 45: | Line 45: | ||
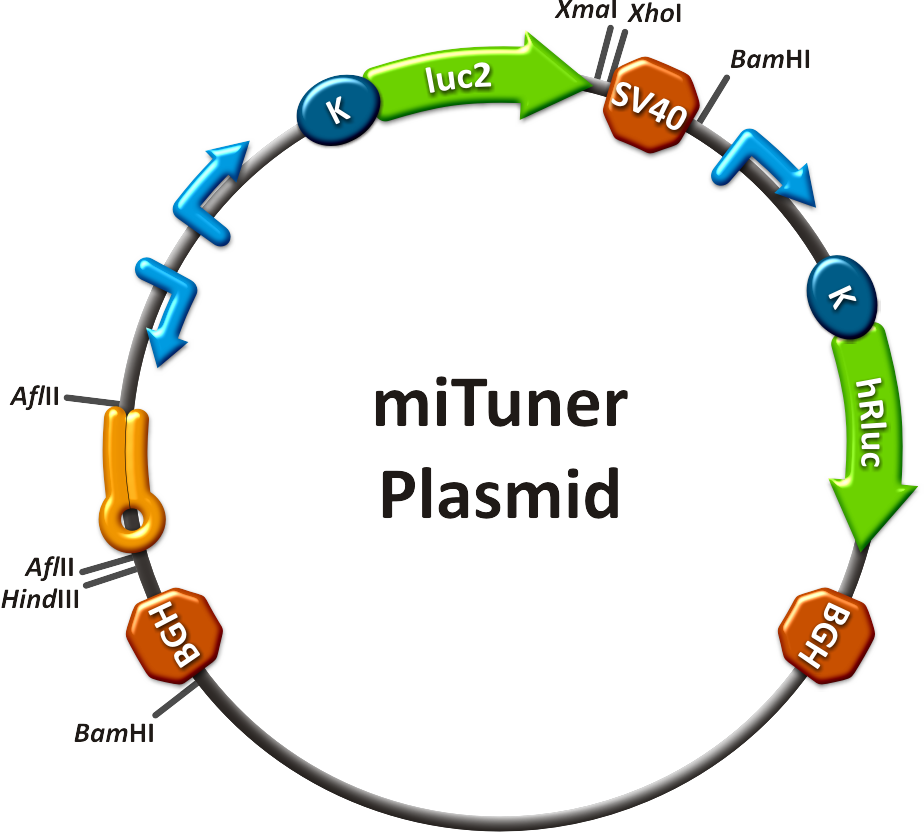
===miTuner: Expression Fine-Tuning by Synthetic miRNAs=== | ===miTuner: Expression Fine-Tuning by Synthetic miRNAs=== | ||
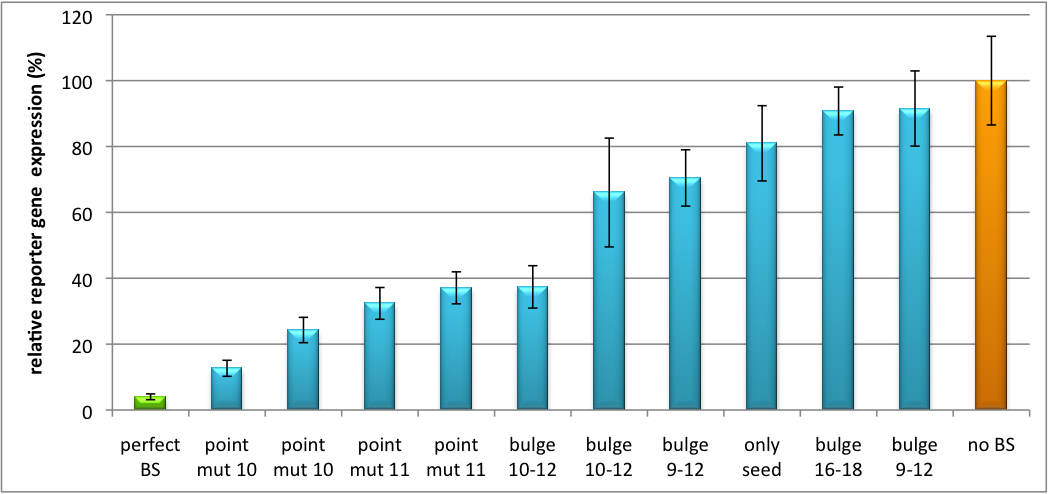
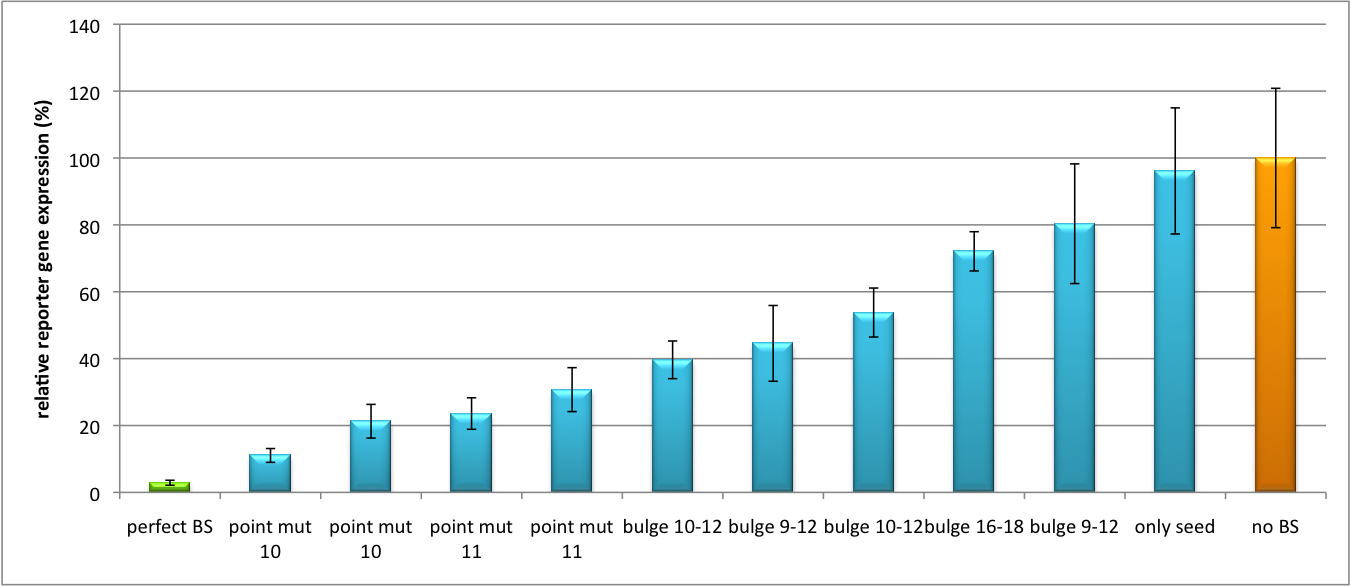
| - | The data shows a precisely tuned expression from almost 0% to 100% (Fig. | + | The data shows a precisely tuned expression from almost 0% to 100% (Fig. 2, Fig. 3). Lowest expression refers to complete knockdown through fusion of perfect binding sites (always green bar on the left hand side of the figures) to the reporter gene. Expression from a construct without binding sites is set as 100% (always orange column on the right hand side of the figures). In presence of the specific shRNA miR, gene expression was mediated to various levels through interactions with the different imperfect binding sites. Whereas, when an unspecific shRNA miR was expressed, gene expression remained unaffected (see raw data below). This reference shows that the binding sites were correctly designed, since they seem to interact specifically with a referring shRNA miR. |
| - | [[Image:Haat_H1HD2010.jpg|thumb|center|600px|'''Figure | + | [[Image:Haat_H1HD2010.jpg|thumb|center|600px|'''Figure 2: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_H1.''' Gene expression quantified via dual luciferase assay for constructs containing different imperfect binding sites for shhAAT.]] |
| - | Figure | + | Figure 2 shows the results of Dual-Luciferase measurements of the miTuner plasmid with binding sites against shhAAT behind firefly luciferase. The highest knockdown can be achieved by using a perfect binding site. Single mutations outside the seed region at position 11, 12 or 10-12 lead to knockdown between 10% and 60% compared to unregulated expression. Bulges close to the seed region or changes in the seed region itself lead to very low downregulation. Having only the seed region as a target for the miRNA also leads to a less efficient knockdown compared with binding sites containing flanking regions. |
| - | [[Image:Haat_U6HD2010.jpg|thumb|center|600px|'''Figure | + | [[Image:Haat_U6HD2010.jpg|thumb|center|600px|'''Figure 3: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_U6.''' Gene expression quantified via dual luciferase assay for constructs containing different imperfect binding sites for shhAAT.]] |
| - | Figure | + | Figure 3 shows the same assay using binding sites against shhAAT within the pBS U6 vector. The results are overall similar, with changes in or directly adjacent to the seed region having the highest impact on knockdown efficiency. The measurement uses the same binding sites as the one conducted in pBS U6, just having a H1 promoter instead of U6 promoter. |
| - | [[Image:PsiCheck.png|thumb|center|600px|'''Figure | + | [[Image:PsiCheck.png|thumb|center|600px|'''Figure 4: Tuning of gene expression through different imperfect miR122 binding sites in psiCHECK-2.''' Construct was transfected into HeLa cells together with an plasmid expressing miR122. Control without binding site was used for normalization.]] |
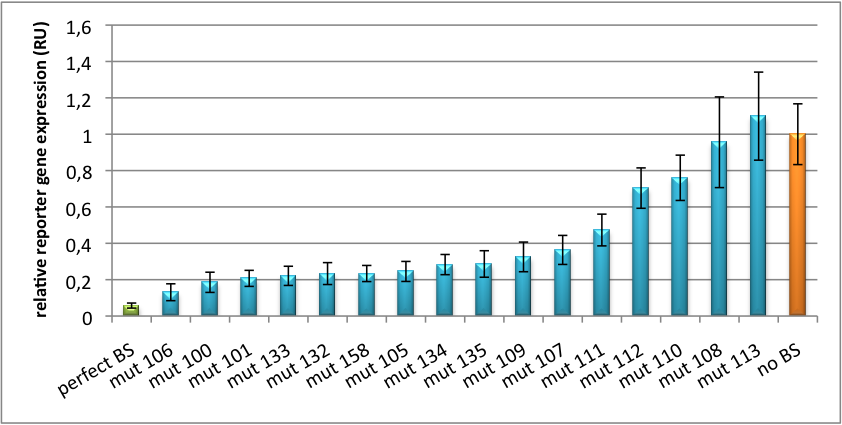
| - | We further analyzed binding sites derived from miR122 in the dual luciferase vector PsiCheck2 as can be seen in | + | We further analyzed binding sites derived from miR122 in the dual luciferase vector PsiCheck2 as can be seen in figure 4. Here we tested sixteen mutated binding sites in order to observe minute fine-tuning between one binding site and the next. Mutated Binding sites 123, 133, 134, 135 and 158 contain 4bp-bulges (non-paired regions) that don not seem to diminish knockdown efficiency much. 107 contains one binding site, while 134 and 135 contain two binding sites for the same miRNA and show a stronger knockdown than 107. |
===Off-Targeting Using Endogenous miRNA=== | ===Off-Targeting Using Endogenous miRNA=== | ||
| Line 64: | Line 64: | ||
To enable Off-Targeting, the GOI expressed on miTuner can be tagged with a miRNA binding site specific for one or a combination of endogenous miRNA of the tissue that is to be excluded from gene expression. | To enable Off-Targeting, the GOI expressed on miTuner can be tagged with a miRNA binding site specific for one or a combination of endogenous miRNA of the tissue that is to be excluded from gene expression. | ||
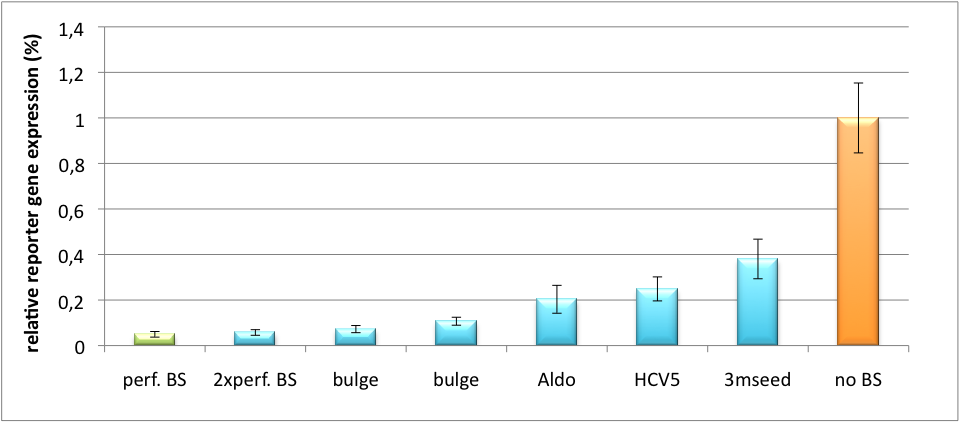
| - | [[Image:HuH Offpng.png|thumb|center|500px|'''Figure | + | [[Image:HuH Offpng.png|thumb|center|500px|'''Figure 5: Knockdown of reporter gene expression due to endogenous miR122 that interferes with binding sites.''' Construct transfected to HuH cells to off-target those.]] |
===On-Targeting Using Endogenous miRNA=== | ===On-Targeting Using Endogenous miRNA=== | ||
Revision as of 18:00, 27 October 2010

|
|
||
 "
"