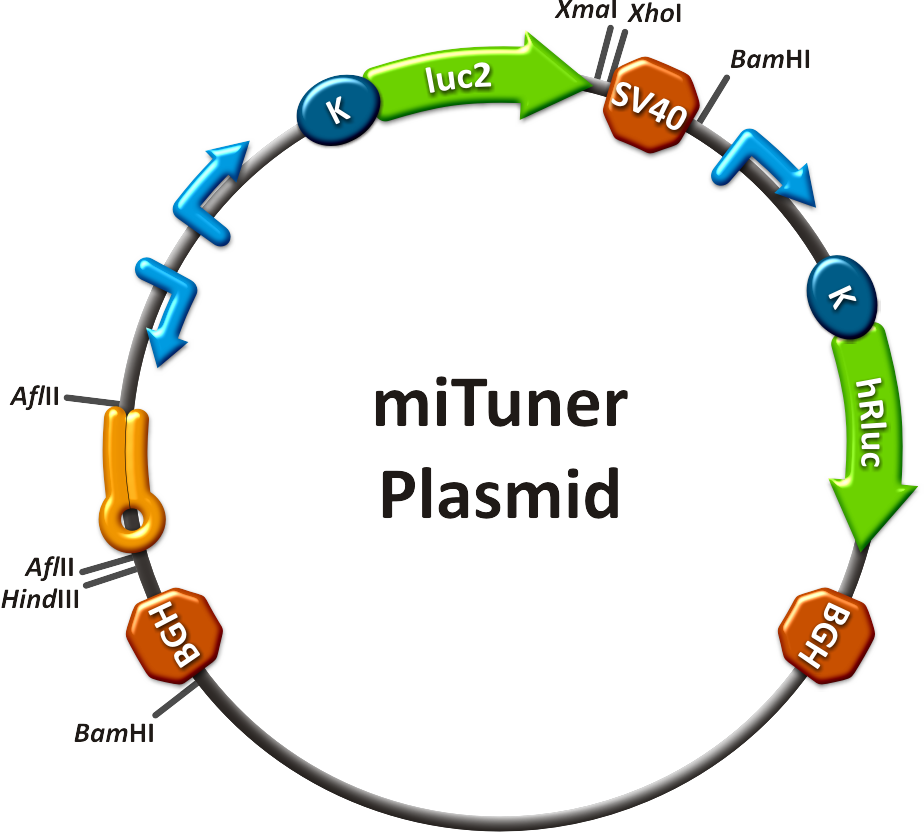
Team:Heidelberg/Project/miRNA Kit
From 2010.igem.org
Laura Nadine (Talk | contribs) (→Results) |
Laura Nadine (Talk | contribs) (→miTuner: Expression Fine-Tuning by Synthetic miRNAs) |
||
| Line 53: | Line 53: | ||
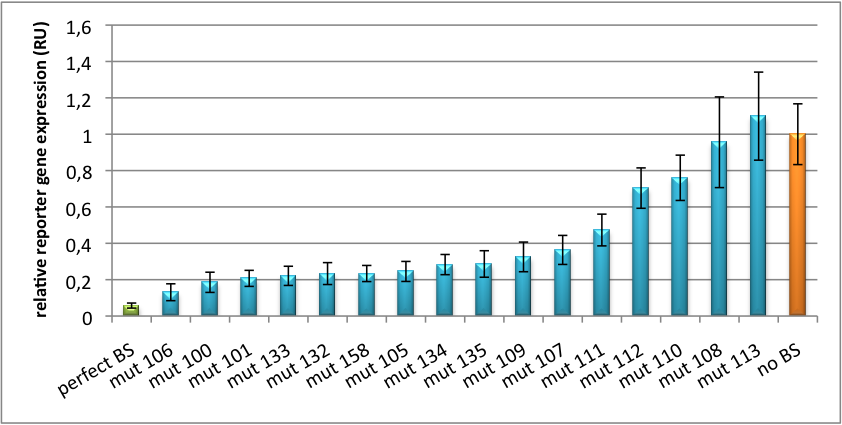
[[Image:Haat_U6HD2010.jpg|thumb|center|600px|'''Figure 2: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_U6.''' Gene expression quantified via dual luciferase assay for constructs containing different imperfect binding sites for shhAAT.]] | [[Image:Haat_U6HD2010.jpg|thumb|center|600px|'''Figure 2: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_U6.''' Gene expression quantified via dual luciferase assay for constructs containing different imperfect binding sites for shhAAT.]] | ||
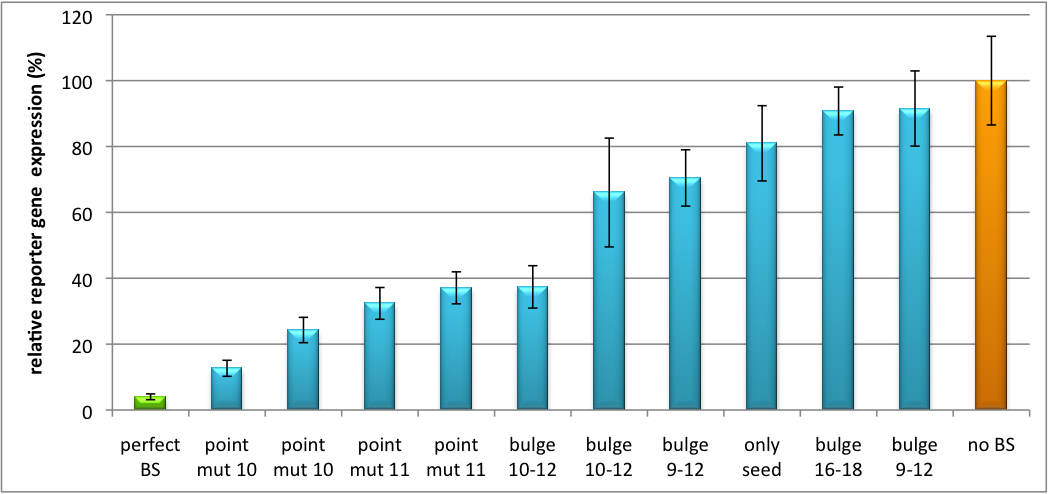
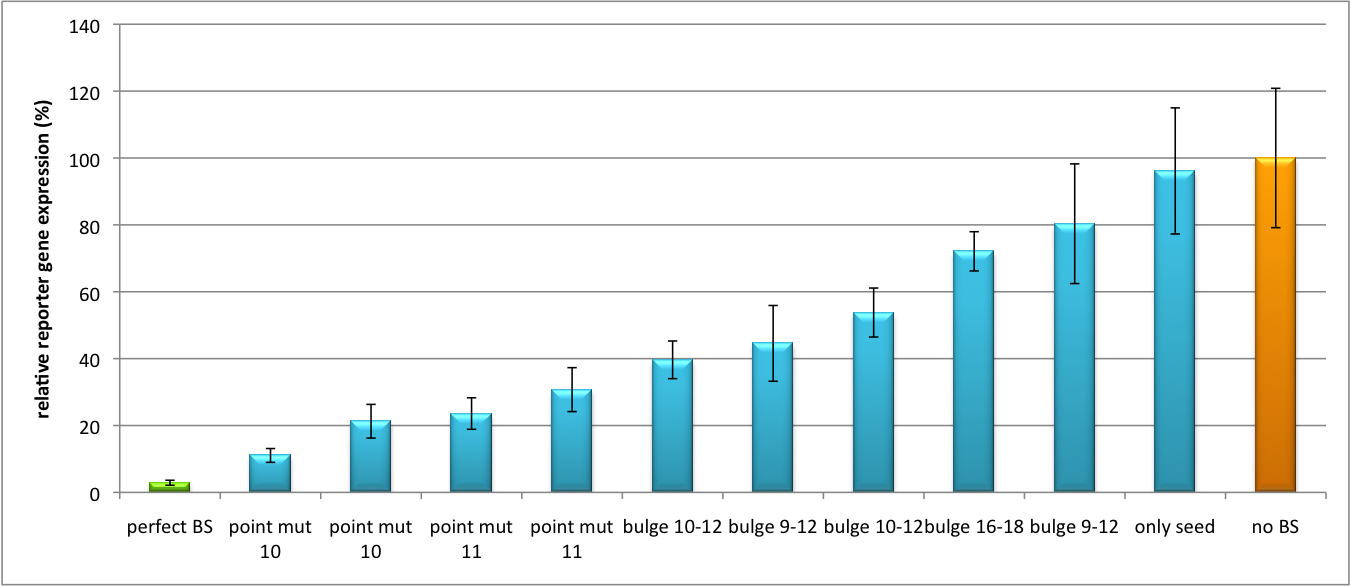
| - | Figure 2 shows the same assay using binding sites against shhAAT within the pBS U6 vector. | + | Figure 2 shows the same assay using binding sites against shhAAT within the pBS U6 vector. The results are overall similar, with changes in or directly adjacent to the seed region having the highest impact on knockdown efficiency. The measurement uses the same binding sites as the one conducted in pBS U6, just having a H1 promoter instead of U6 promoter. |
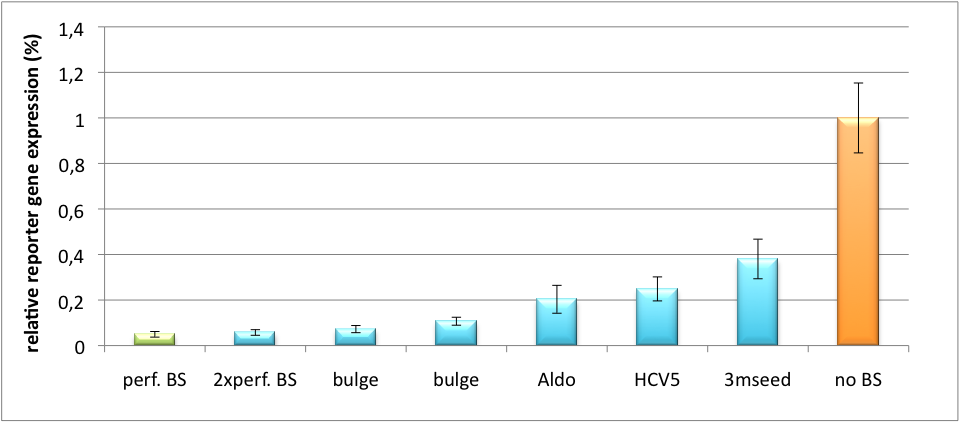
[[Image:PsiCheck.png|thumb|center|600px|'''Figure 3: Tuning of gene expression through different imperfect miR122 binding sites in psiCHECK-2.''' Construct was transfected into HeLa cells together with an plasmid expressing miR122. Control without binding site was used for normalization.]] | [[Image:PsiCheck.png|thumb|center|600px|'''Figure 3: Tuning of gene expression through different imperfect miR122 binding sites in psiCHECK-2.''' Construct was transfected into HeLa cells together with an plasmid expressing miR122. Control without binding site was used for normalization.]] | ||
Revision as of 17:13, 27 October 2010

|
|
||
 "
"