Team:Heidelberg/Project/miRNA Kit
From 2010.igem.org
Laura Nadine (Talk | contribs) (→Discussion) |
Laura Nadine (Talk | contribs) |
||
| Line 13: | Line 13: | ||
<div class="t1">Synthetic miRNA Kit</div> | <div class="t1">Synthetic miRNA Kit</div> | ||
| - | <h4>miTuner - a kit for microRNA based gene expression tuning in mammalian cells</h4> | + | <center><h4>miTuner - a kit for microRNA based gene expression tuning in mammalian cells</h4></center> |
<br/> | <br/> | ||
<center><i>With the synthetic miRNA kit, we provide a comprehensive mean | <center><i>With the synthetic miRNA kit, we provide a comprehensive mean | ||
| Line 28: | Line 28: | ||
'''On-targeting''' is based on the expression of the GOI from a promoter containing a Tet Operon that negatively regulates gene expression in the presence of a Tet Repressor ('''figure 1c'''). If the Tet Repressor is under control of perfect binding sites for endogenous miRNAs , it will be downregulated in the target tissue, releasing the promoter and enabling specific GOI expression. | '''On-targeting''' is based on the expression of the GOI from a promoter containing a Tet Operon that negatively regulates gene expression in the presence of a Tet Repressor ('''figure 1c'''). If the Tet Repressor is under control of perfect binding sites for endogenous miRNAs , it will be downregulated in the target tissue, releasing the promoter and enabling specific GOI expression. | ||
| - | |||
==Introduction== | ==Introduction== | ||
| Line 44: | Line 43: | ||
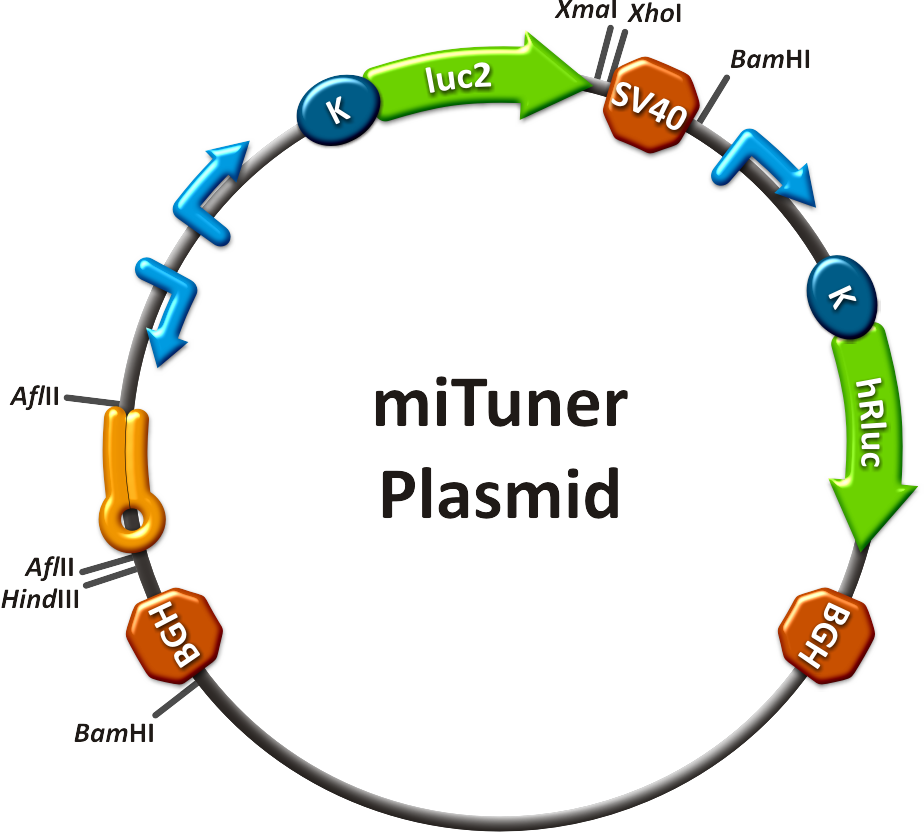
All gene regulatory constructs for tuning, Off- or On-targeting can easily be assembled using '''BBB standard cloning''' from our miRNA Kit [https://2010.igem.org/Team:Heidelberg/Parts parts]. After successful cloning, the constructs can be transfected onto a cell line of choice or transferred into a virus backbone for [https://2010.igem.org/Team:Heidelberg/Project/Mouse_Infection ''in vivo''] experiments. For our '''prove of principle''', we used firefly luciferase normalized to ''Renilla'' luciferase on miTuner to characterize knockdown efficiencies of different binding sites and show Off- and On-targeting by mouse infection carried by an AAV virus. | All gene regulatory constructs for tuning, Off- or On-targeting can easily be assembled using '''BBB standard cloning''' from our miRNA Kit [https://2010.igem.org/Team:Heidelberg/Parts parts]. After successful cloning, the constructs can be transfected onto a cell line of choice or transferred into a virus backbone for [https://2010.igem.org/Team:Heidelberg/Project/Mouse_Infection ''in vivo''] experiments. For our '''prove of principle''', we used firefly luciferase normalized to ''Renilla'' luciferase on miTuner to characterize knockdown efficiencies of different binding sites and show Off- and On-targeting by mouse infection carried by an AAV virus. | ||
| - | ===miTuner: Expression | + | ===miTuner: Expression Fine-Tuning by Synthetic miRNAs=== |
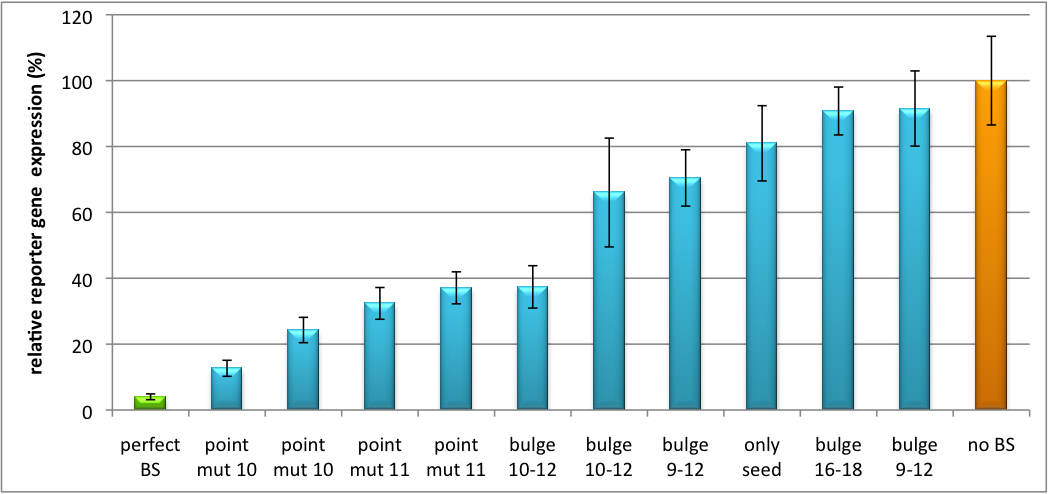
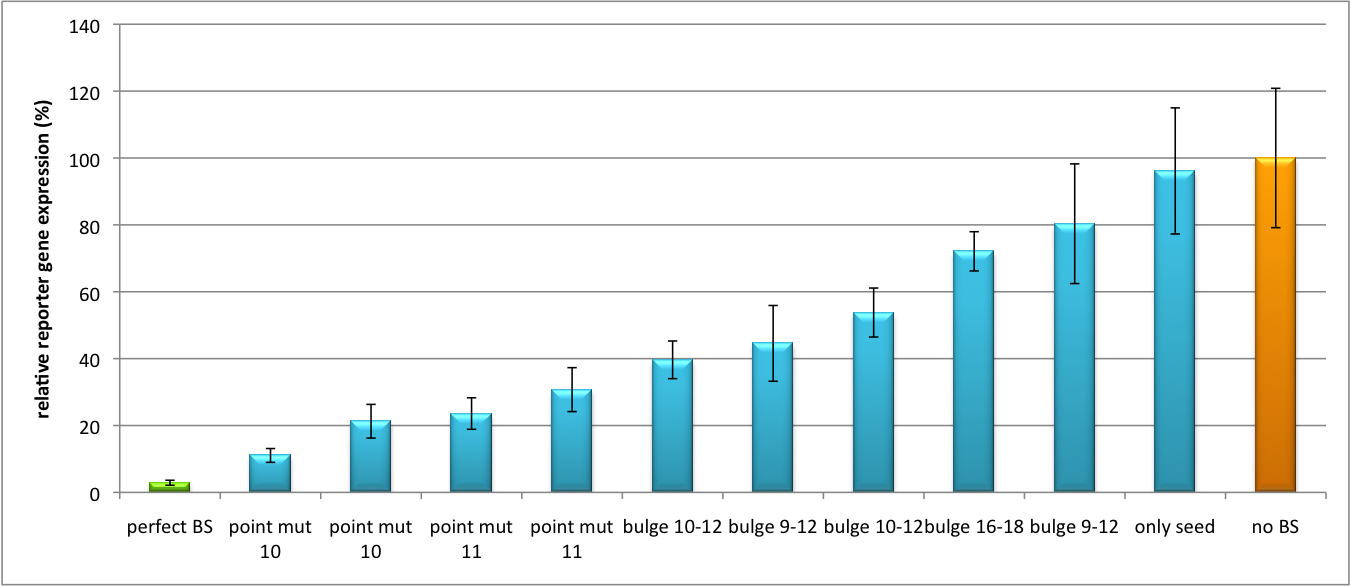
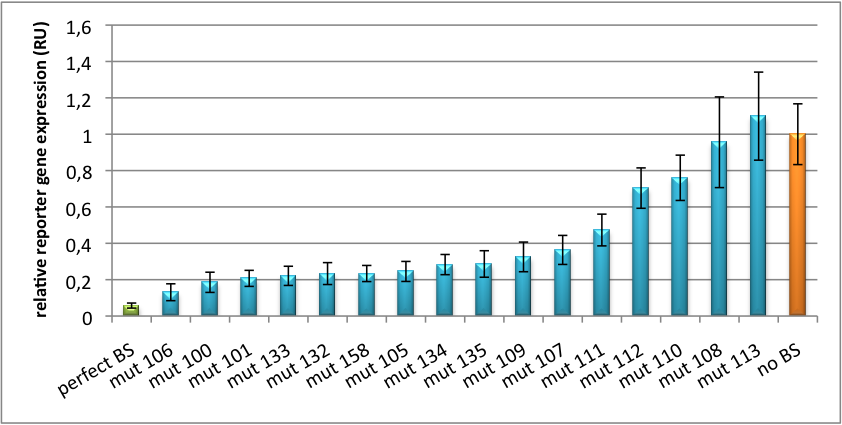
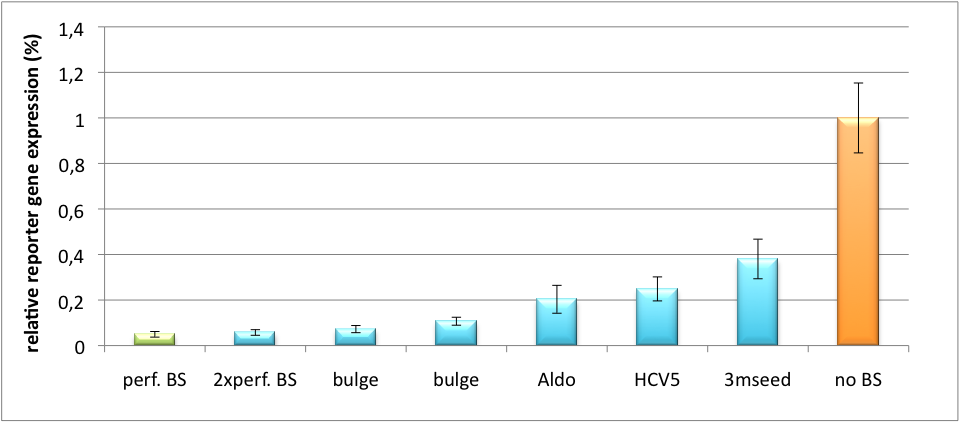
| - | The data shows a precisely tuned expression from almost 0% to 100% (Fig. 1, Fig. 2). Lowest expression refers to complete knockdown through fusion of perfect binding sites (always green bar on the left hand side of the figures) to the reporter gene. | + | The data shows a precisely tuned expression from almost 0% to 100% (Fig. 1, Fig. 2). Lowest expression refers to complete knockdown through fusion of perfect binding sites (always green bar on the left hand side of the figures) to the reporter gene. Expression from a construct without binding sites is set as 100% (always orange column on the right hand side of the figures). In presence of the specific shRNA miR, gene expression was mediated to various levels through interactions with the different imperfect binding sites. Whereas, when an unspecific shRNA miR was expressed, gene expression remained unaffected (see raw data below). This reference shows that the binding sites were correctly designed, since they seem to interact specifically with a referring shRNA miR. |
[[Image:Haat_H1HD2010.jpg|thumb|center|600px|'''Figure 1: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_H1.''' Gene expression quantified via dual luciferase assay for constructs containing different imperfect binding sites for shhAAT.]] | [[Image:Haat_H1HD2010.jpg|thumb|center|600px|'''Figure 1: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_H1.''' Gene expression quantified via dual luciferase assay for constructs containing different imperfect binding sites for shhAAT.]] | ||
Revision as of 16:22, 27 October 2010

|
|
||
 "
"