Team:Heidelberg/Project/miRNA Kit
From 2010.igem.org
(→Results) |
(→Results) |
||
| Line 34: | Line 34: | ||
==Results== | ==Results== | ||
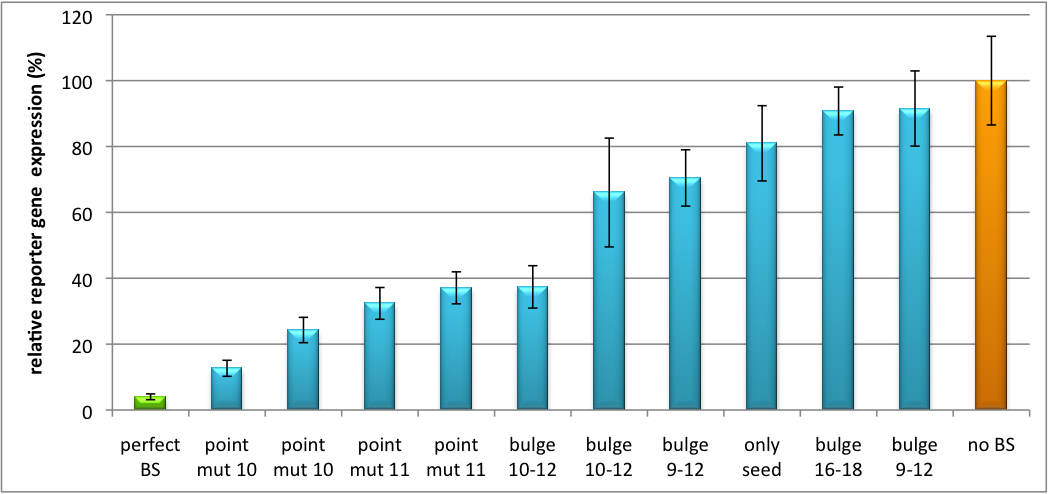
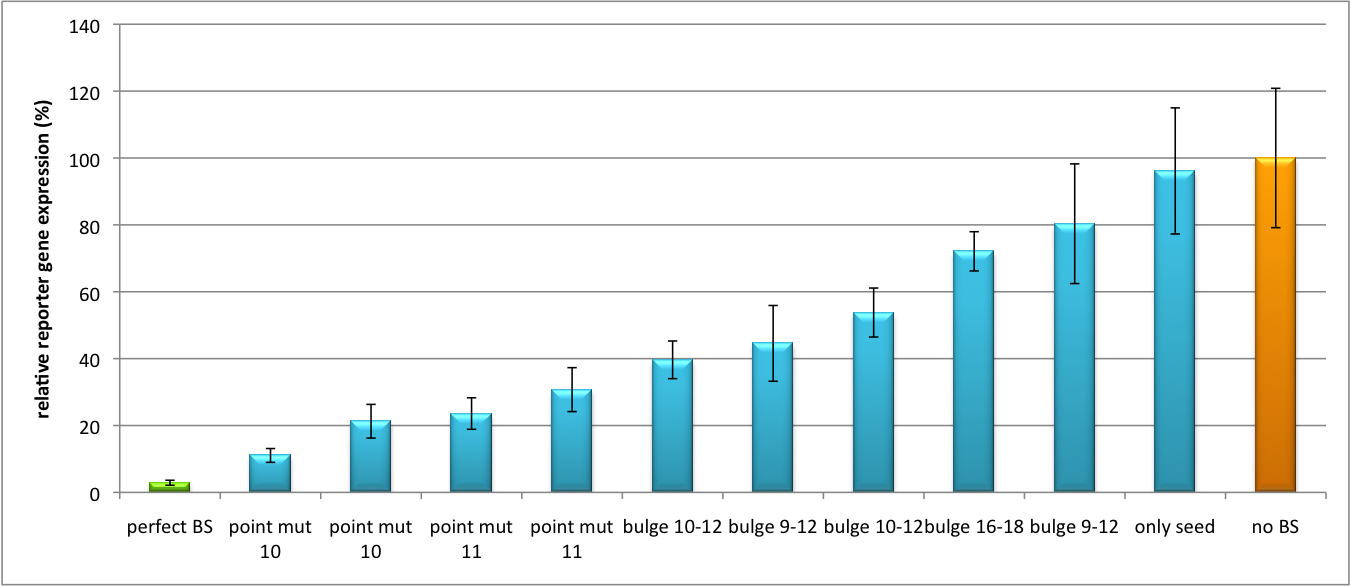
| - | Regulation of gene expression can be achieved by fusing miRNA binding sites right behind a GOI. In case a referring shRNA miR is expressed, the GOI is knocked down. Strength of regulation thereby depends on binding site properties. We are able to tune gene expression linearly over a broad range. This is a first proof of principle for various miRNA-mRNA interaction <i>in vitro</i>. Therefore, we transfected [https://2010.igem.org/Team:Heidelberg/Notebook/Material#Cell_lines HeLa cells] in principle with our [http://partsregistry.org/Part:BBa_K337036 pSMB_miTuner Plasmid HD3]. It turned out, that there was no obvious effect of different binding sites on reporter gene expression (data not shown). We assume that the RSV driving the shRNA miR is too weak for tight regulation of the referring binding site behind the GOI. Only if a sufficient amount of shRNA miR binds to its target, translation is significantly repressed. Thus, we expressed the shRNA miR from a separate plasmid which was always co-transfected with the original tuning construct. The reporter genes - i. e. Luc2 and hRluc - were also expressed from separate plasmids to get a reference as well as a transfection control. Then, we conducted a [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Dual_Luciferase_Assay Dual Luciferase Assay] for quantification of gene expression. The data preciously shows a tuned expression from almost 0% to 100% (Fig. 1, Fig. 2). Lowest expression refers to complete knockdown through fusion of perfect binding sites (always green bar on the left hand side of the figures) to the reporter gene. 100% means ordinary expression from a construct without binding sites (always orange column on the right hand side of the figures). In presence of the specific shRNA miR, gene expression was mediated to various levels through interactions with the different imperfect binding sites. Whereas, when an unspecific shRNA miR was expressed, gene expression remained unaffected (see raw data below). The latter aspect reveals, that the binding sites were correctly designed, since they seem to interact specifically with a referring shRNA miR. The constructs were tested in two different backbones: pBS_U6 and pBS_H1. Both are in viral context, meaning that they contain inverted terminal repeats (ITRs). The constructs can be packed into the capsid of an adeno-associated virus (AAV). Those constructs we also chose for [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Virus_Production virus production] to infect cells even more efficiently as compared to transfections. Because of the significant data, we decided to inject the viruses into mice to see the tuning effect also <i>[https://2010.igem.org/Team:Heidelberg/Project/Mouse_Infection in vivo]</i>. The pBS_H1 construct should be preferred for mice injections since the expressed shRNA miR against human alpha-1-antitrypsine (shhAAT) is cytotoxic in higher concentrations. The pBS_H1 backbone leads to moderate expression ranges, still obviously showing the tuning effect. | + | Regulation of gene expression can be achieved by fusing miRNA binding sites right behind a GOI. In case a referring shRNA miR is expressed, the GOI is knocked down. Strength of regulation thereby depends on binding site properties. We are able to tune gene expression linearly over a broad range. This is a first proof of principle for various miRNA-mRNA interaction <i>in vitro</i>. Therefore, we transfected [https://2010.igem.org/Team:Heidelberg/Notebook/Material#Cell_lines HeLa cells] in principle with our [http://partsregistry.org/Part:BBa_K337036 pSMB_miTuner Plasmid HD3]. It turned out, that there was no obvious effect of different binding sites on reporter gene expression (data not shown). We assume that the RSV driving the shRNA miR is too weak for tight regulation of the referring binding site behind the GOI. Only if a sufficient amount of shRNA miR binds to its target, translation is significantly repressed. Thus, we expressed the shRNA miR from a separate plasmid which was always co-transfected with the original tuning construct. The reporter genes - i. e. Luc2 and hRluc - were also expressed from separate plasmids to get a reference as well as a transfection control. Then, we conducted a [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Dual_Luciferase_Assay Dual Luciferase Assay] for quantification of gene expression. The data preciously shows a tuned expression from almost 0% to 100% (Fig. 1, Fig. 2). Lowest expression refers to complete knockdown through fusion of perfect binding sites (always green bar on the left hand side of the figures) to the reporter gene. 100% means ordinary expression from a construct without binding sites (always orange column on the right hand side of the figures). In presence of the specific shRNA miR, gene expression was mediated to various levels through interactions with the different imperfect binding sites. Whereas, when an unspecific shRNA miR was expressed, gene expression remained unaffected (see raw data below). The latter aspect reveals, that the binding sites were correctly designed, since they seem to interact specifically with a referring shRNA miR. The constructs were tested in two different backbones: pBS_U6 and pBS_H1. Both are in viral context, meaning that they contain inverted terminal repeats (ITRs). The constructs can be packed into the capsid of an adeno-associated virus (AAV). Those constructs we also chose for [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Virus_Production virus production] to infect cells even more efficiently as compared to transfections. Because of the significant data, we decided to inject the viruses into mice to see the tuning effect also <i>[https://2010.igem.org/Team:Heidelberg/Project/Mouse_Infection in vivo]</i>. The pBS_H1 construct should be preferred for mice injections since the expressed synthetic shRNA miR against human alpha-1-antitrypsine (shhAAT) is cytotoxic in higher concentrations. The pBS_H1 backbone leads to moderate expression ranges, still obviously showing the tuning effect. |
[[Image:Haat_H1HD2010.jpg|thumb|center|600px|'''Figure 1: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_H1.''' Gene expression quantified via dual luciferase assay for constructs containing different imperfect binding sites for shhAAT.]] | [[Image:Haat_H1HD2010.jpg|thumb|center|600px|'''Figure 1: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_H1.''' Gene expression quantified via dual luciferase assay for constructs containing different imperfect binding sites for shhAAT.]] | ||
[[Image:Haat_U6HD2010.jpg|thumb|center|600px|'''Figure 2: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_U6.''' Gene expression quantified via dual luciferase assay for constructs containing different imperfect binding sites for shhAAT.]] | [[Image:Haat_U6HD2010.jpg|thumb|center|600px|'''Figure 2: Tuning of gene expression through different imperfect shRNA miR binding sites in pBS_U6.''' Gene expression quantified via dual luciferase assay for constructs containing different imperfect binding sites for shhAAT.]] | ||
| - | Strikingly, the order of constructs in terms of knockdown for the imperfect binding sites is similar. M4, M5 and M6 always show strong knockdown, whereas M9, M10 and M11 show only loose down-regulation. Consulting the binding site sequences, the weak knockdown can be addressed to bulges in the supplementary region or complete lack of the 3' region of the binding site. Still high strength could be maintained due to only single nucleotide exchanges in the central region of the binding site. | + | Strikingly, the order of constructs in terms of knockdown for the imperfect binding sites is similar. M4, M5 and M6 always show strong knockdown, whereas M9, M10 and M11 show only loose down-regulation. Consulting the binding site sequences, the weak knockdown can be addressed to bulges in the supplementary region or to complete lack of the 3' region of the binding site. Still high strength could be maintained due to only single nucleotide exchanges in the central region of the binding site. |
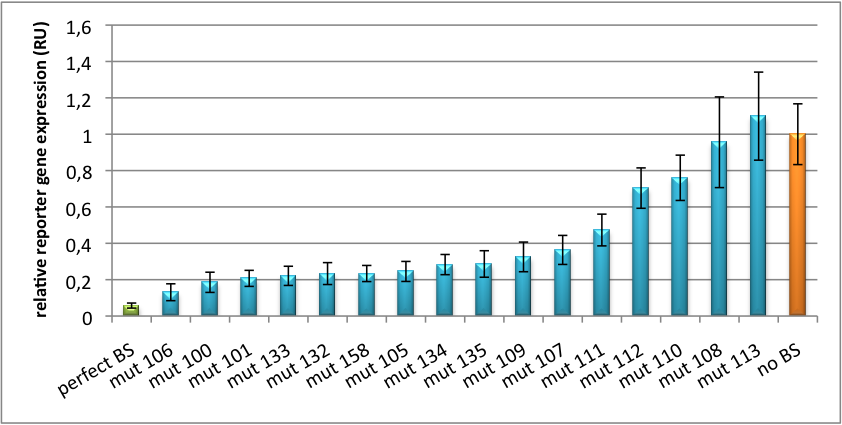
| + | The principle of smooth regulation was also demonstrated for miR122, a microRNA that is exclusively upregulated in hepatic cells. Referring binding sites were cloned into psiCHECK-2 backbone (Promega) and due to sequence mutations different Luciferase levels were detected again (Fig. 3). | ||
| + | [[Image:PsiCheck.png|thumb|center|600px|'''Figure 3: Tuning of gene expression through different imperfect miR122 binding sites in psiCHECK-2.''' Construct was transfected into HeLa cells together with an plasmid expressing miR122. Control without binding site was used for normalization.]] | ||
| + | We can state: the gene expression tuning with miBricks works for synthetic as well as endogenous miRNA. | ||
<!--After creating a binding site library and testing the miRNA-binding site interaction <i>in vitro</i>, we were able to compute an [https://2010.igem.org/Team:Heidelberg/Modeling/miGUI <i>in silico</i> model] based on a machine learning approach to predict knockdown efficiencies. A more detailed description of the different binding sites, we characterized can be found in our [https://2010.igem.org/Team:Heidelberg/Project/miMeasure measurements] page. | <!--After creating a binding site library and testing the miRNA-binding site interaction <i>in vitro</i>, we were able to compute an [https://2010.igem.org/Team:Heidelberg/Modeling/miGUI <i>in silico</i> model] based on a machine learning approach to predict knockdown efficiencies. A more detailed description of the different binding sites, we characterized can be found in our [https://2010.igem.org/Team:Heidelberg/Project/miMeasure measurements] page. | ||
Revision as of 08:27, 27 October 2010

|
|
||
 "
"