Team:Freiburg Bioware/NoteBook/Labjournal/Cellculture
From 2010.igem.org
(→30.9 Seeding AAV293 for production of vectors with mVenus missing beta globin) |
|||
| (232 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Freiburg_Bioware/css}} | {{:Team:Freiburg_Bioware/css}} | ||
| - | |||
{{:Team:Freiburg_Bioware/Head}} | {{:Team:Freiburg_Bioware/Head}} | ||
| + | {{:Team:Freiburg_Bioware/menu_notebook}} | ||
<!-- Freiburg_bioware --> | <!-- Freiburg_bioware --> | ||
| Line 20: | Line 20: | ||
<li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/September">September part 1 (labday 107 - 123)</a></li> | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/September">September part 1 (labday 107 - 123)</a></li> | ||
<li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/September2">September part 2 (labday 124 - 135)</a></li> | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/September2">September part 2 (labday 124 - 135)</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/October">October part 1 (labday 136 - | + | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/October">October part 1 (labday 136 - 149 )</a></li> |
| - | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/October2">October part 2 (labday | + | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/October2">October part 2 (labday 150 - 166 )</a></li> |
| - | + | ||
<li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/November">November (labday 167 - 170 )</a></li> | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/November">November (labday 167 - 170 )</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/Cellculture">Cellculture </a></li> | + | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/Cellculture">Cellculture</a></li> |
</ul> | </ul> | ||
</div> | </div> | ||
| Line 55: | Line 54: | ||
<br /> | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| - | AAV 293 and HT1080 Cells have been splitted and plated out on 10 cm dishes for transfektion. | + | |
| - | + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | |
| - | After harvesting | + | <tr> |
| - | 2,5µl of this cell suspension have been mixed with 47.5µl of trypan blue. | + | <th width="100">Days</th> |
| + | <th width="100">14.6</th> | ||
| + | <th width="100">15.6</th> | ||
| + | <th width="100">16.6</th> | ||
| + | <th width="100">17.6</th> | ||
| + | <th width="100">18.6</th> | ||
| + | <th width="100">19.6</th> | ||
| + | <th width="100">20.6</th> | ||
| + | <th width="100">21.6</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>-</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest + Transduction</td> | ||
| + | <td></td> | ||
| + | <td>Fluorescence microscopy</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | AAV 293 and HT1080 Cells have been splitted and plated out on 10 cm dishes for transfektion. After harvesting the cells according to the standard protocol the cell-pellets were resuspendiated in 15ml of DYT medium. | ||
| + | 2,5µl of this cell suspension have been mixed with 47.5µl of trypan blue and the cells were counted by using the Neubauer-Meteringchamber. | ||
We counted 12,5x 10^6 cells/ml for the AAV293 cell line and 10x10^6 cells /ml for the HT1080 cell line. | We counted 12,5x 10^6 cells/ml for the AAV293 cell line and 10x10^6 cells /ml for the HT1080 cell line. | ||
| - | + | 1ml and 1,5 ml of the AAV 293 cells have been seeded on four dishes with 250µl of the cell suspension. | |
| - | + | Note that that the date is wrong on the plates and the flasks! Cells have been already plated out on the 14th. of June. | |
| - | + | ||
| - | We also seated two flasks one for each cell line. Therefor we used 1ml of the HT1080 cell suspension and | + | We also seated two flasks one for each cell line. Therefor we used 1ml of the HT1080 cell suspension and 2ml of the the AAV293 cells. |
<br /> | <br /> | ||
1) | 1) | ||
| Line 90: | Line 111: | ||
Adrian tried to examine the precipation of the CaCl2+ DNA Clusters. We have to optimize the pH of our 2xHBS at the moment it is 11.12. | Adrian tried to examine the precipation of the CaCl2+ DNA Clusters. We have to optimize the pH of our 2xHBS at the moment it is 11.12. | ||
<br /> | <br /> | ||
| - | The viral stocks | + | The viral stocks were harvested according to standard protocol. |
<br> | <br> | ||
<b>The results</b><br /> | <b>The results</b><br /> | ||
| Line 106: | Line 127: | ||
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
| - | We want to repeat the last transfection | + | We want to repeat the last transfection and obtain a vsibly higher GOI (mVenus) expression. |
<br /> | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">19.6</th> | ||
| + | <th width="100">20.6</th> | ||
| + | <th width="100">20.6</th> | ||
| + | <th width="100">21.6</th> | ||
| + | <th width="100">22.6</th> | ||
| + | <th width="100">23.6</th> | ||
| + | <th width="100">24.6</th> | ||
| + | <th width="100">25.6</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>-</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest + Transduction</td> | ||
| + | <td></td> | ||
| + | <td>Fluorescence microscopy</td> | ||
| + | </tr> | ||
| + | </table> | ||
Six transfections were performed according to the standart protocol:<br> | Six transfections were performed according to the standart protocol:<br> | ||
Three 10 cm cellcluture dishes with 293 cells were transfected with 10 µg DNA (3,33 µg each plasmid) and the other three 10 cm cellculture dishes with 30 µg DNA (10 µg DNA each plasmid). | Three 10 cm cellcluture dishes with 293 cells were transfected with 10 µg DNA (3,33 µg each plasmid) and the other three 10 cm cellculture dishes with 30 µg DNA (10 µg DNA each plasmid). | ||
| Line 133: | Line 178: | ||
The Transduction efficency is still quite low! We have to optimize the stratagene protocol! | The Transduction efficency is still quite low! We have to optimize the stratagene protocol! | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;">30.6 Examination of different Transduction methods </p>=== | + | ===<p style="font-size:17px; background-color:#00dd77;">30.6 Examination of different Transduction methods</p>=== |
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
| - | We want to | + | We want to check whether the amount of our produced AAV2s has an effect on the transduction rate. |
| + | Instead of 500µl, 1000µl of the AAV-stock were pipetted into each well. | ||
<br /><br /> | <br /><br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">30.6</th> | ||
| + | <th width="100">1.7</th> | ||
| + | <th width="100">2.7</th> | ||
| + | <th width="100">3.7</th> | ||
| + | <th width="100">4.7</th> | ||
| + | <th width="100">5.7</th> | ||
| + | <th width="100">6.7</th> | ||
| + | <th width="100">7.7</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>-</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest + Transduction</td> | ||
| + | <td></td> | ||
| + | <td>Fluorescence microscopy</td> | ||
| + | </tr> | ||
| + | </table> | ||
1. Plate I(A is up) | 1. Plate I(A is up) | ||
| Line 151: | Line 221: | ||
{| border="1" | {| border="1" | ||
| - | | 1000 µl of Viral Stock DROPPED || align="right" |1000 µl of Viral Stock gently resuspended|| align="right" |1500 µl of Viral Stock | + | | 1000 µl of Viral Stock DROPPED || align="right" |1000 µl of Viral Stock gently resuspended|| align="right" |1500 µl of Viral Stock rough resuspending |
|- | |- | ||
| 1000 µl of Viral Stock DROPPED || align="right" | 1000 µl of Viral Stock gently resuspended|| align="right" | no Transduction | | 1000 µl of Viral Stock DROPPED || align="right" | 1000 µl of Viral Stock gently resuspended|| align="right" | no Transduction | ||
|- | |- | ||
|} | |} | ||
| - | |||
<br> | <br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<b>The results</b><br /> | <b>The results</b><br /> | ||
| - | All cells | + | All cells died probably because we forgot to wash the cells with PBS before adding fresh medium. It was not possible to take pictures because the dead cells were all clumped. |
Looks like we should not transduce 3x10^5 cells with 1000 µl (or more) of our viral solution because almost all cells died in all approaches and the medium indicator was yellow. | Looks like we should not transduce 3x10^5 cells with 1000 µl (or more) of our viral solution because almost all cells died in all approaches and the medium indicator was yellow. | ||
| - | So we can not say if resuspendig the cells with the viral soultion is a useful alteration of the standard protocol. | + | So we can not say if resuspendig the cells with the viral soultion is a useful alteration of the standard protocol. |
<br> | <br> | ||
| Line 188: | Line 240: | ||
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
| - | We want to check if the amount of plasmids is the reason for the low transduction efficiency. | + | We want to check if the amount of plasmids is the reason for the low transduction efficiency. Unfortunately the flow cytometry (FACS) is not available at the moment so the results can only be checked via fuorescence microscopy. |
<br /><br /> | <br /><br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| - | Transfection with P41 was performed according standart protocol. | + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > |
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">3.7</th> | ||
| + | <th width="100">4.7</th> | ||
| + | <th width="100">5.7</th> | ||
| + | <th width="100">6.7</th> | ||
| + | <th width="100">7.7</th> | ||
| + | <th width="100">8.7</th> | ||
| + | <th width="100">9.7</th> | ||
| + | <th width="100">10.7</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>-</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest + Transduction</td> | ||
| + | <td></td> | ||
| + | <td>Fluorescence microscopy</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | Transfection with P41 (pAAV_iGEM-MCS_mVenus) was performed according standart protocol. | ||
Two transfections where carried out with 10µg (3,33 µg each plasmid) DNA and the other two with 20µg DNA (6,66 µg each plasmid).<br /> | Two transfections where carried out with 10µg (3,33 µg each plasmid) DNA and the other two with 20µg DNA (6,66 µg each plasmid).<br /> | ||
We want to investigate which of the following approaches yields a better transduction efficiency. | We want to investigate which of the following approaches yields a better transduction efficiency. | ||
| Line 206: | Line 282: | ||
* 3 cycles of freezing and thawing | * 3 cycles of freezing and thawing | ||
<br /> | <br /> | ||
| - | * used plasmids: 10µg, 20µg | + | * used plasmids: 10µg, 20µg GOI-plasmid (mVenus) |
| - | * we have 10µg and 20µg from standart protocol => | + | * we have 10µg and 20µg from standart protocol => standard virus |
* we have 10µg and 20µg from standart protocol pellet => Pellet | * we have 10µg and 20µg from standart protocol pellet => Pellet | ||
* we have 10µg and 20µg from standart protocol suspension => Super | * we have 10µg and 20µg from standart protocol suspension => Super | ||
| Line 253: | Line 329: | ||
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
| - | We want to check if the mVenus expression | + | We want to check if the mVenus expression and transduction efficiency can be improved with higher amounts of DNA. |
<br /><br /> | <br /><br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">4.7</th> | ||
| + | <th width="100">5.7</th> | ||
| + | <th width="100">6.7</th> | ||
| + | <th width="100">7.7</th> | ||
| + | <th width="100">8.7</th> | ||
| + | <th width="100">9.7</th> | ||
| + | <th width="100">10.7</th> | ||
| + | <th width="100">11.7</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>-</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest + Transduction</td> | ||
| + | <td></td> | ||
| + | <td>FACS</td> | ||
| + | </tr> | ||
| + | </table> | ||
The Transfection was performed with 10, 18 and 24 µg per plasmid (mVenus, pHelper and R/C). | The Transfection was performed with 10, 18 and 24 µg per plasmid (mVenus, pHelper and R/C). | ||
<br /> | <br /> | ||
| - | + | The virus harvest was performed according to standard protocol: | |
* 3 Plates got transduced | * 3 Plates got transduced | ||
1. Plate I(A is up) completely 10 µg | 1. Plate I(A is up) completely 10 µg | ||
{| border="1" | {| border="1" | ||
| - | | 300µl | + | | 300µl resuspended|| align="right" |300µl resuspended|| align="right" |450µl resuspended |
|- | |- | ||
| - | | 600µl | + | | 600µl resuspended|| align="right" |600µl resuspended|| align="right" | control |
|- | |- | ||
|} | |} | ||
| Line 272: | Line 372: | ||
2. Plate II(A is up)completely 18 µg | 2. Plate II(A is up)completely 18 µg | ||
{| border="1" | {| border="1" | ||
| - | | 300µl | + | | 300µl resuspended|| align="right" |300µl resuspended|| align="right" |450µl resuspended |
|- | |- | ||
| - | | 600µl | + | | 600µl resuspended|| align="right" |600µl resuspended|| align="right" | control |
|- | |- | ||
|} | |} | ||
| Line 281: | Line 381: | ||
3. Plate III(A is up) completely 24 µg | 3. Plate III(A is up) completely 24 µg | ||
{| border="1" | {| border="1" | ||
| - | | 300µl | + | | 300µl resuspended|| align="right" |300µl resuspended|| align="right" |450µl resuspended |
|- | |- | ||
| - | | 600µl | + | | 600µl resuspended|| align="right" |600µl resuspended|| align="right" | control |
|- | |- | ||
|} | |} | ||
| Line 319: | Line 419: | ||
17= Plate 3 B2 : 20,8% " from 75,7% <br> | 17= Plate 3 B2 : 20,8% " from 75,7% <br> | ||
18= Plate 3 B3 : 0,07% " from 84,8% <br> | 18= Plate 3 B3 : 0,07% " from 84,8% <br> | ||
| - | |||
| - | |||
| - | |||
| Line 336: | Line 433: | ||
<br /> | <br /> | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;">14.7 Seeding AAV293 for TKGMK constructs | + | ===<p style="font-size:17px; background-color:#00dd77;">14.7 Seeding AAV293 for TKGMK constructs </p>=== |
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
| Line 342: | Line 439: | ||
<br /><br /> | <br /><br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">14.7</th> | ||
| + | <th width="100">15.7</th> | ||
| + | <th width="100">16.7</th> | ||
| + | <th width="100">17.7</th> | ||
| + | <th width="100">18.7</th> | ||
| + | <th width="100">19.7</th> | ||
| + | <th width="100">20.7</th> | ||
| + | <th width="100">21.7</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>-</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest + Transduction</td> | ||
| + | <td></td> | ||
| + | <td>FACS</td> | ||
| + | </tr> | ||
| + | </table> | ||
30µg Plasmid were used (pHelper + pAAV_RC+ pAAV_RFC25_mgmk_TK30)<br> | 30µg Plasmid were used (pHelper + pAAV_RC+ pAAV_RFC25_mgmk_TK30)<br> | ||
VIRUS no GOI :pHelper + pAAV_RC<br> | VIRUS no GOI :pHelper + pAAV_RC<br> | ||
| Line 351: | Line 472: | ||
<br /> | <br /> | ||
<b>The results</b><br /> | <b>The results</b><br /> | ||
| - | Unfortunately the stocks never were used. | + | Unfortunately the stocks never were used (exception: see following experiment, 7. august) because later on we noticed that the TKGMK plasmids were not in line with the RFC. |
<br /> | <br /> | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;">7.8 Seeding AAV293 for TKGMK and mVenus stocks </p>=== | + | ===<p style="font-size:17px; background-color:#00dd77;">7.8 Seeding AAV293 for TKGMK and mVenus stocks</p>=== |
| - | <b>The motivation</b | + | <b>The motivation</b> |
| - | + | After proofing the successfull transduction of tumor cells with the reportergen mVenus the next step is to kill the cells with a prodrug approach. The prodrug of our choice is the guanosine anolog ganciclovir, once activated ganciclovir gets integrated into the growing dna chain which results in termination of dna synthesis and cell death. We transduce the tumor cell line HT1080 with viral particles packed with the thymidine kinase guanosine mono phosphate kinase fusion protein (TKGMK) as gene of interest and the incubation with ganciclovir should kill the cells.<br /> | |
<br /> | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">7.8</th> | ||
| + | <th width="100">8.8</th> | ||
| + | <th width="100">9.8</th> | ||
| + | <th width="100">10.8</th> | ||
| + | <th width="100">11.8</th> | ||
| + | <th width="100">12.8</th> | ||
| + | <th width="100">13.8</th> | ||
| + | <th width="100">14.8</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>-</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest + Transduction</td> | ||
| + | <td></td> | ||
| + | <td>FACS and microscopy</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br /> | ||
{| border="1" | {| border="1" | ||
| Plate || align="right" |pAAV_RC/µg|| align="right" |pHelper/µg|| align="right" |GOI/µg | | Plate || align="right" |pAAV_RC/µg|| align="right" |pHelper/µg|| align="right" |GOI/µg | ||
| Line 369: | Line 515: | ||
| 2|| align="right" |6,6 || align="right" | 3,3|| align="right" | no GOI | | 2|| align="right" |6,6 || align="right" | 3,3|| align="right" | no GOI | ||
|- | |- | ||
| - | | 3|| align="right" |3,3 || align="right" | 3,3|| align="right" | 3,3 TK_GMK clone1 | + | | 3|| align="right" |3,3 || align="right" | 3,3|| align="right" | 3,3 TK_GMK clone1 (P54) |
|- | |- | ||
| - | | 4|| align="right" |3,3 || align="right" | 3,3|| align="right" | 3,3 TK_GMK clone1 | + | | 4|| align="right" |3,3 || align="right" | 3,3|| align="right" | 3,3 TK_GMK clone1 (P54) |
|- | |- | ||
| - | | 5|| align="right" |3,3 || align="right" | 3,3|| align="right" | 3,3 TK_GMK clone2 | + | | 5|| align="right" |3,3 || align="right" | 3,3|| align="right" | 3,3 TK_GMK clone2 (P55) |
|- | |- | ||
| 6|| align="right" |3,3 || align="right" | 3,3|| align="right" | 3,3 YFP | | 6|| align="right" |3,3 || align="right" | 3,3|| align="right" | 3,3 YFP | ||
| Line 386: | Line 532: | ||
|- | |- | ||
|} | |} | ||
| - | The transduction was performed according to standard protocol. | + | <br />The transduction was performed according to standard protocol. |
| - | + | Plate 1:<br /> | |
<table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
| Line 411: | Line 557: | ||
</table> | </table> | ||
<br /> | <br /> | ||
| - | + | Plate 2:<br /> | |
<table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
| Line 434: | Line 580: | ||
</table> | </table> | ||
<br /> | <br /> | ||
| - | + | Plate 3:<br /> | |
<table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
| Line 457: | Line 603: | ||
</table> | </table> | ||
<br /> | <br /> | ||
| - | + | Plate 4:<br /> | |
<table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
| Line 480: | Line 626: | ||
</table> | </table> | ||
<br /> | <br /> | ||
| - | + | Plate 5:<br /> | |
<table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
| Line 503: | Line 649: | ||
</table> | </table> | ||
<br /> | <br /> | ||
| - | + | Plate 6:<br /> | |
<table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
| Line 526: | Line 672: | ||
</table> | </table> | ||
<br /> | <br /> | ||
| - | + | Plate 7:<br /> | |
<table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
| Line 549: | Line 695: | ||
</table> | </table> | ||
<br /> | <br /> | ||
| - | + | Plate 8:<br /> | |
<table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
| Line 590: | Line 736: | ||
As you can see, the TKGMK cell killing mechanism works! The samples with targeted mVenus constructs couldnt be checked because the FACS machine was not working. The fluorescence was checked with microscopy, the transduction efficiency is still low. | As you can see, the TKGMK cell killing mechanism works! The samples with targeted mVenus constructs couldnt be checked because the FACS machine was not working. The fluorescence was checked with microscopy, the transduction efficiency is still low. | ||
| - | + | ===<p style="font-size:17px; background-color:#00dd77;"><b>14.8 Seeding different amounts of AAV293 for transfection</b></p>=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;"><b>14.8 Seeding different amounts of AAV293 for transfection </b></p>=== | + | |
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
We want to know which confluence of AAV293 cells is optimal for AAV production.<br /> | We want to know which confluence of AAV293 cells is optimal for AAV production.<br /> | ||
<br /> | <br /> | ||
<b>The plan</b> | <b>The plan</b> | ||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">14.8</th> | ||
| + | <th width="100">15.8</th> | ||
| + | <th width="100">16.8</th> | ||
| + | <th width="100">17.8</th> | ||
| + | <th width="100">18.8</th> | ||
| + | <th width="100">19.8</th> | ||
| + | <th width="100">20.8</th> | ||
| + | <th width="100">21.8</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>-</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest + Transduction</td> | ||
| + | <td></td> | ||
| + | <td>FACS</td> | ||
| + | </tr> | ||
| + | </table> | ||
<br /> | <br /> | ||
We seed different amounts of cells in 10 cm<sup>2</sup>, and check the highest titer.<br /> | We seed different amounts of cells in 10 cm<sup>2</sup>, and check the highest titer.<br /> | ||
<br /> | <br /> | ||
{| border="1" | {| border="1" | ||
| - | | Plate || align="right" |amount of cells | + | | Plate || align="right" |amount of seeded AAV-293 cells |
|- | |- | ||
| 1|| align="right" |100 000 | | 1|| align="right" |100 000 | ||
| Line 654: | Line 793: | ||
|} | |} | ||
| - | The cells were transfected with 40µg | + | The cells were transfected with 40µg standard CMV_YFP, 3,3µg Rep/Cap and 3,3µg pHelper. |
Transduction was performed according to standard protocol. | Transduction was performed according to standard protocol. | ||
| Line 771: | Line 910: | ||
</table> | </table> | ||
<b>The results</b><br /> | <b>The results</b><br /> | ||
| + | The FACS was still not available but we could easily detect a significant higher YFP expression (70-80%) at the following positions: | ||
| + | * Plate 1: A2 and B2, A3 and B3 | ||
| + | * Plate 2: A2 and B2, A3 and B3 | ||
| + | Therefore we conclude that we have to seed between 100000 and 500000 cells for optimal virus production precedure. | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;">17.8 transduction with viral stocks 20 µg and 40 µg</p>=== | + | ===<p style="font-size:17px; background-color:#00dd77;">17.8 transduction with viral stocks 20 µg and 40 µg GOI-plasmid</p>=== |
| - | + | <b>The Motivation</b><br /> | |
| + | We want to FACS our stocks to get valid data for further investigation of optimal virus assembly. Therefore created stocks with 20 and 40 µg CMV_mVenus. | ||
| + | <br /><b>The Plan</b><br /> | ||
| + | <br /> | ||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">17.8</th> | ||
| + | <th width="100">18.8</th> | ||
| + | <th width="100">19.8</th> | ||
| + | <th width="100">20.8</th> | ||
| + | <th width="100">21.8</th> | ||
| + | <th width="100">22.8</th> | ||
| + | <th width="100">23.8</th> | ||
| + | <th width="100">24.8</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>-</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest + Transduction</td> | ||
| + | <td></td> | ||
| + | <td>FACS</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br /> | ||
<li>1. plate 50.000 cells, YFP | <li>1. plate 50.000 cells, YFP | ||
{| align=right | {| align=right | ||
| Line 834: | Line 1,005: | ||
|- | |- | ||
|} | |} | ||
| - | < | + | <b>The Results</b><br /> |
| - | + | Although we were told that the FACS would be available in time it wasn't. So the cells could unfortunately not be checked for their YFP expression =(. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
===<p style="font-size:17px; background-color:#00dd77;">16.8 Seeding AAV293 for transfection with new R/C</p>=== | ===<p style="font-size:17px; background-color:#00dd77;">16.8 Seeding AAV293 for transfection with new R/C</p>=== | ||
<b>The motivation</b> <br /> | <b>The motivation</b> <br /> | ||
| - | We want examine the functionality of different R/C constructs and the new eGFP gene of interest. This transfection was performed with new cells which were thawed. | + | We want examine the functionality of different R/C constructs and the new eGFP gene of interest. This transfection was performed with new AAV-293 cells which were thawed. |
<b>The plan</b> <br /> | <b>The plan</b> <br /> | ||
| - | + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | |
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">16.8</th> | ||
| + | <th width="100">17.8</th> | ||
| + | <th width="100">18.8</th> | ||
| + | <th width="100">19.8</th> | ||
| + | <th width="100">20.8</th> | ||
| + | <th width="100">21.8</th> | ||
| + | <th width="100">22.8</th> | ||
| + | <th width="100">23.8</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>-</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest + Transduction</td> | ||
| + | <td></td> | ||
| + | <td>FACS</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | * 2x10<sup>6</sup> cells were seeded | ||
* used plasmids: | * used plasmids: | ||
**P50 c= 429 ng/µl (RC) | **P50 c= 429 ng/µl (RC) | ||
| Line 873: | Line 1,050: | ||
The transfection was performed according to standard protocol. | The transfection was performed according to standard protocol. | ||
| - | * <b> 1. plate:</b> Lipo-Transfection | + | * <b> 1. plate:</b> Lipo-Transfection => named Lipo |
| - | * <b> 2. plate:</b> Lipo-Transfection | + | * <b> 2. plate:</b> Lipo-Transfection => named Lipo |
| - | * <b> 3. plate:</b> 3,3 µg (RC), 3,3 µg (pHelper), 3,3 µg (YFP) | + | * <b> 3. plate:</b> 3,3 µg (RC), 3,3 µg (pHelper), 3,3 µg (YFP) => was not used for transduction |
| - | * <b> 4. plate:</b> 10 µg (RC), 10 µg (pHelper), 3,3 µg (YFP) | + | * <b> 4. plate:</b> 10 µg (RC), 10 µg (pHelper), 3,3 µg (YFP) => named CaCl<sub>2</sub> |
| - | * <b> 5. plate:</b> 10 µg (RC mutant P158a), 10 µg (pHelper), 3,3 µg (YFP) | + | * <b> 5. plate:</b> 10 µg (RC mutant P158a), 10 µg (pHelper), 3,3 µg (YFP)=> CaCl<sub>2</sub> 10 µg RC mutant P158a |
| - | * <b> 6. plate:</b> 10 µg (RC mutant P158b), 10 µg (pHelper), 3,3 µg (YFP) | + | * <b> 6. plate:</b> 10 µg (RC mutant P158b), 10 µg (pHelper), 3,3 µg (YFP)=> CaCl<sub>2</sub> 10 µg RC mutant P158b |
| - | * <b> 7. plate:</b> 3,3 µg (RC mutant P158a), 3,3 µg (pHelper), 3,3 µg (YFP) | + | * <b> 7. plate:</b> 3,3 µg (RC mutant P158a), 3,3 µg (pHelper), 3,3 µg (YFP)=> CaCl<sub>2</sub> 3,3 µg RC mutant P158a |
| - | * <b> 8. plate:</b> 3,3 µg (RC mutant P158b), 3,3 µg (pHelper), 3,3 µg (YFP) | + | * <b> 8. plate:</b> 3,3 µg (RC mutant P158b), 3,3 µg (pHelper), 3,3 µg (YFP)=> CaCl<sub>2</sub> 3,3 µg RC mutant P158b |
| - | * <b> 9. plate:</b> 10 µg (RC), 10 µg (pHelper), 3,3 µg (eGFP) | + | * <b> 9. plate:</b> 10 µg (RC), 10 µg (pHelper), 3,3 µg (eGFP)=> CaCl<sub>2</sub> eGFP 3,3 µg |
| - | * <b> 10. plate:</b> 10 µg (RC), 10 µg (pHelper), 10 µg (eGFP) | + | * <b> 10. plate:</b> 10 µg (RC), 10 µg (pHelper), 10 µg (eGFP)=> CaCl<sub>2</sub> eGFP 10 µg |
| - | * <b> 11. plate:</b> 10 µg (RC), 10 µg (pHelper), 20 µg (eGFP) | + | * <b> 11. plate:</b> 10 µg (RC), 10 µg (pHelper), 20 µg (eGFP)=> CaCl<sub>2</sub> eGFP 20 µg |
| - | * <b> 12. plate:</b> 10 µg (RC), 10 µg (pHelper), 40 µg (eGFP) | + | * <b> 12. plate:</b> 10 µg (RC), 10 µg (pHelper), 40 µg (eGFP)=> CaCl<sub>2</sub> eGFP 40 µg |
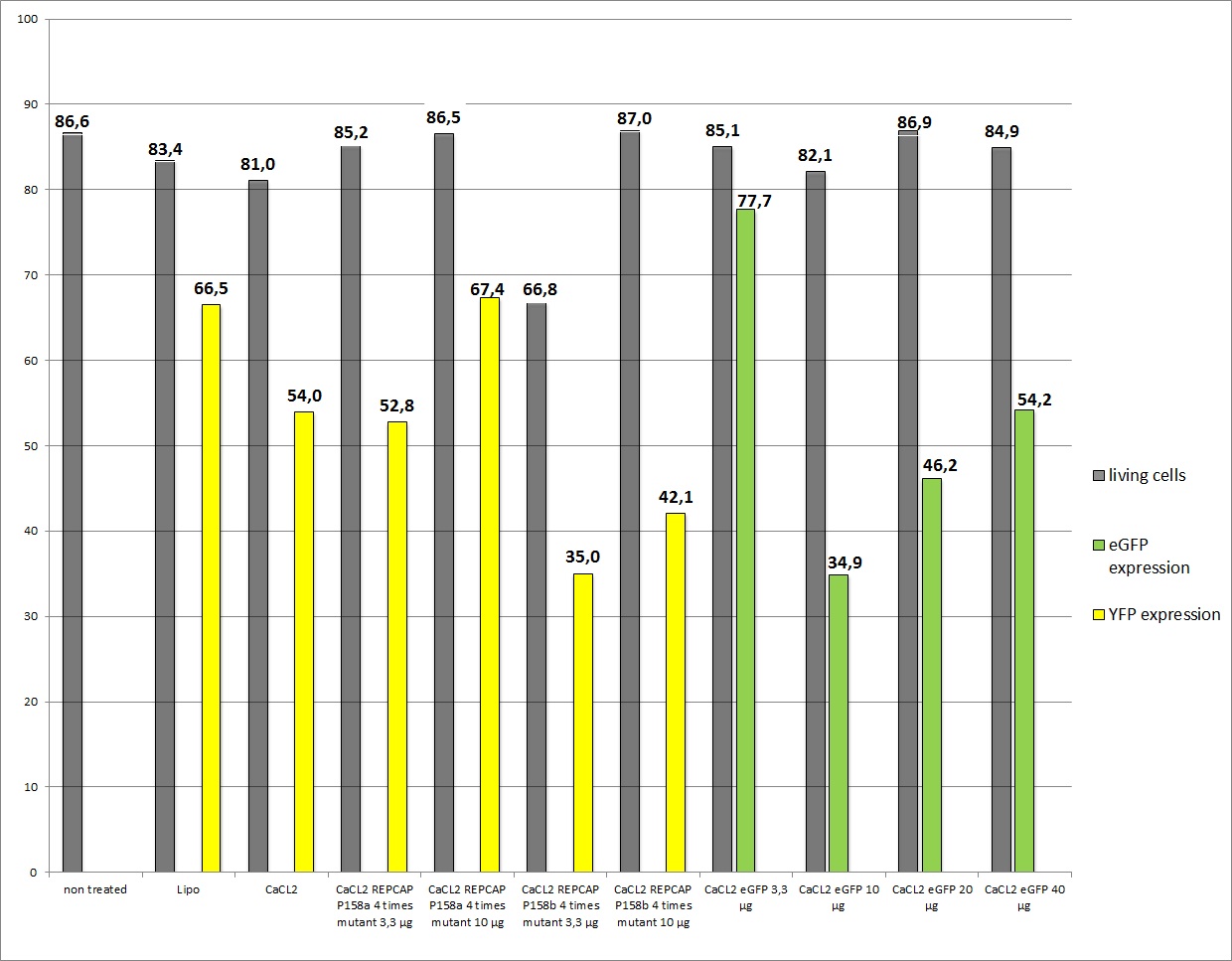
<b>The results</b> <br /> | <b>The results</b> <br /> | ||
| Line 890: | Line 1,067: | ||
[[Image:Freiburg10 23.8.2010 FACS eGFP Lipofection 200.000.jpg|800px]] | [[Image:Freiburg10 23.8.2010 FACS eGFP Lipofection 200.000.jpg|800px]] | ||
| - | As you can see the eGFP constructs very well. And even the transduction efficiency of the standard R/C is | + | As you can see the eGFP constructs worked very well. And even the transduction efficiency of the standard R/C is ok, the conclusion is that in previous transfections the AAV293 cells were to confluent so their transfection capability. The confluence of the cells is crucial! It also seems that our used amounts of plasmids had no decisive effect on the YFP expression because of the obviously variating results. A final conclusion is still due. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
===<p style="font-size:17px; background-color:#00dd77;">19.8 Seeding AAv293 for transfection with different HBS buffers </p>=== | ===<p style="font-size:17px; background-color:#00dd77;">19.8 Seeding AAv293 for transfection with different HBS buffers </p>=== | ||
| Line 910: | Line 1,075: | ||
We want to try different 2xHBS Buffers (pH) and their influence on transfection efficiency!<br /><br /> | We want to try different 2xHBS Buffers (pH) and their influence on transfection efficiency!<br /><br /> | ||
<b>The Plan</b><br /> | <b>The Plan</b><br /> | ||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">19.8</th> | ||
| + | <th width="100">20.8</th> | ||
| + | <th width="100">21.8</th> | ||
| + | <th width="100">22.8</th> | ||
| + | <th width="100">23.8</th> | ||
| + | <th width="100">24.8</th> | ||
| + | <th width="100">25.8</th> | ||
| + | <th width="100">26.8</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>-</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest + Transduction</td> | ||
| + | <td></td> | ||
| + | <td>FACS</td> | ||
| + | </tr> | ||
| + | </table> | ||
We try 5 different 2xHBS buffers: | We try 5 different 2xHBS buffers: | ||
| Line 921: | Line 1,110: | ||
<br /> | <br /> | ||
| - | Used Plasmids | + | Used Plasmids: 10µg of each plasmid |
<ul> | <ul> | ||
<li>GOI: mVENUS => P262, conc:1148ng/µl; used amount: 8,71µl</li> | <li>GOI: mVENUS => P262, conc:1148ng/µl; used amount: 8,71µl</li> | ||
| Line 928: | Line 1,117: | ||
<li>RepCap => 10.8: 1348ng/µl; used amount: 7,41µl</li> | <li>RepCap => 10.8: 1348ng/µl; used amount: 7,41µl</li> | ||
</ul> | </ul> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br /> | <br /> | ||
*Transduction of three 6-well plates: | *Transduction of three 6-well plates: | ||
| Line 1,013: | Line 1,190: | ||
*(1)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,06 | *(1)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,06 | ||
*(2)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,08 | *(2)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,08 | ||
| - | *(3)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7, | + | *(3)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,112 |
*(4)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,12 | *(4)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,12 | ||
*(5)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,14 | *(5)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,14 | ||
| - | |||
<br /> | <br /> | ||
<b>the Results</b><br /> | <b>the Results</b><br /> | ||
AAV-Harvesting is at 24.8, same day Transduction and FACS will be done at 26.8 | AAV-Harvesting is at 24.8, same day Transduction and FACS will be done at 26.8 | ||
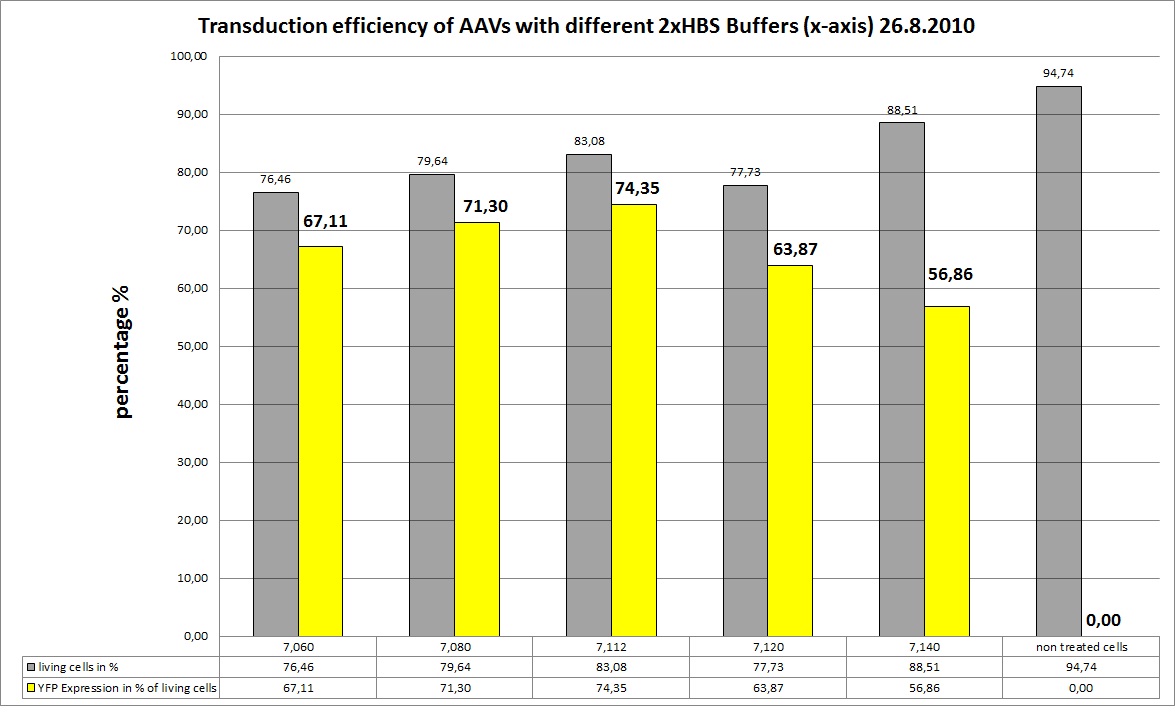
| - | [[Image:Freiburg10 26.8 | + | [[Image:Freiburg10 CC 26.8 different HBS.jpg|800px]] |
| - | As you can see the 2xHBS with pH | + | As you can see the 2xHBS with pH 7.112 performed best, in future experiments this buffer will be used. |
<br /> | <br /> | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;"> | + | ===<p style="font-size:17px; background-color:#00dd77;">22.8: Transfection of AAV293 with different amounts of cells, and two plasmid concentrations</p>=== |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
<br /> | <br /> | ||
| Line 1,065: | Line 1,208: | ||
<br /> | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
| Line 1,115: | Line 1,235: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | The following amount of cells were seeded in 10 cm<sup>2</sup>: | ||
| + | <ol> | ||
| + | <li>100.000</li> | ||
| + | <li>100.000</li> | ||
| + | <li>200.000</li> | ||
| + | <li>200.000</li> | ||
| + | <li>300.000</li> | ||
| + | <li>300.000</li> | ||
| + | <li>400.000</li> | ||
| + | <li>400.000</li> | ||
| + | <li>500.000</li> | ||
| + | <li>500.000</li> | ||
| + | <li>600.000</li> | ||
| + | <li>600.000</li> | ||
| + | <li>700.000</li> | ||
| + | <li>700.000</li> | ||
| + | </ol> | ||
| + | <br /> | ||
| + | 10µg RC P158a four times mutant, 10µg pHelper, 3,3 µg mVenus P162 were pipetted on plates: 2, 4, 6, 8, 10 , 12, 14 | ||
| + | <br /> | ||
| + | 3,3µg RC P158a four times mutant, 3,3µg pHelper, 3,3 µg mVenus P262 were pipetted on plates: 1, 3, 5, 7, 9, 11, 13 | ||
| + | <br /> | ||
| + | |||
| + | |||
<b>The results</b><br /> | <b>The results</b><br /> | ||
| Line 1,120: | Line 1,264: | ||
[[Image:Freiburg10 30.2010 FACS number of Cells and Beas construct.jpg|800px]] | [[Image:Freiburg10 30.2010 FACS number of Cells and Beas construct.jpg|800px]] | ||
<br /> | <br /> | ||
| - | + | <br /> | |
| - | + | There is only one value for each approach but nevertheless the average YFP expression should be higher. | |
| + | We really dont know the reason for the quite low YFP expression. There are also very good news: the reassembled construct (pSB1C3_leftITR_CMV_betaglobin_mVenus_hGH_rightITR) created from the single Biobricks works as "well" as the AAV2 with CMV_mVenus! | ||
<br/> | <br/> | ||
| Line 1,127: | Line 1,272: | ||
<br /> | <br /> | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;">30.8 Seeding AAV293 for | + | ===<p style="font-size:17px; background-color:#00dd77;">30.8 Seeding AAV293 for Transfection of 10 cm<sup>2</sup> plates</p>=== |
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
We want to create ten hypothetical identical viral stocks to estimate the standard deviation. | We want to create ten hypothetical identical viral stocks to estimate the standard deviation. | ||
| - | <br /> | + | <br /><br /> |
| - | <b>The plan</b> | + | <b>The plan</b> |
| - | + | ||
| - | + | ||
<br /> | <br /> | ||
<table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| Line 1,169: | Line 1,312: | ||
</ul> | </ul> | ||
<br /> | <br /> | ||
| - | 0,5 ml of each stock | + | 0,5 ml of each stock be stored at -80°C.<br /> |
<br /> | <br /> | ||
| - | + | 9,5 ml of each stock will be used for Transduction. <br> | |
| - | + | <br> | |
| + | Result: | ||
| + | <br> | ||
| + | [[Image:Freiburg10_Unbenannt.jpg]] | ||
| + | <br> | ||
| + | Standard deviation: about +/- 4,98% YFP expression. All future AAV2 stocks will be pooled if produced with this same plasmids and amounts of plasmids. Unfortunately the YFP expression is still located at around 50%. We would like to have a YFP expression of at least 70% because as soon as we start to test our not yet ready modified AAV2 we suggest the YFP expression will obviously decrease. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">4.9 Seeding cells for checking constructs (GOIs) with/without HGH and beta globin and reassembled as control </p>=== | ||
<br /> | <br /> | ||
| - | <b>The | + | <b>The motivation</b><br /> |
| - | + | We want to check the functionality of these genes of interest | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
<table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| Line 1,290: | Line 1,357: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | < | + | <ul> |
| + | <li>P269 pSB1C3_lITR_CMV_beta-globin_mVenus_HGH_rITR = 325 ng/µl => 9,66 µl </li> | ||
| + | <li>P270 pSB1C3_lITR_CMV_beta-globin_mVenus_HGH_rITR = 561 ng/µl => 7,26 µl</li> | ||
| + | <li>P377 pSB1C3_lITR_CMV_mVenus_HGH_rITR = 325,04 ng/µl => 30,76 µl </li> | ||
| + | <li>P378 pSB1C3_lITR_CMV_beta-globin_mVenus_rITR = 561 ng/µl => 17,8µl </li> | ||
| + | </ul> | ||
| + | Additionally P50: pAAV_RC R/C (26,45 µl) and pHELPER (14,61µl) pipetted to each approach. <br /> | ||
| + | <br /> | ||
<br /> | <br /> | ||
<b>The results</b><br /> | <b>The results</b><br /> | ||
| Line 1,298: | Line 1,372: | ||
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
| - | + | We want to check the optimal ganciclovir concentration, we use the TK/GMK Stocks (10 µg from each plasmid) from 24.8 with Buffer pH 7,10 and the pH12 stock. | |
| + | <br /> | ||
<br /> | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| - | |||
| - | |||
| - | |||
<table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
| Line 1,332: | Line 1,404: | ||
<br /> | <br /> | ||
<b>the results</b> | <b>the results</b> | ||
| + | <br /> | ||
<br /> | <br /> | ||
[[Image:Freiburg10 CC 6.9 Checking Ganciclovir concentrations-CHART.jpg|800px]] | [[Image:Freiburg10 CC 6.9 Checking Ganciclovir concentrations-CHART.jpg|800px]] | ||
<br /> | <br /> | ||
| - | |||
<br /> | <br /> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br /> | <br /> | ||
| + | As expected the highest concentration of ganciclovir worked the best but it is remarkable that there is no significant difference between the first three ganciclovir amounts. The most remarkable note is, that in future experiments the incubation time has to be extended to 3 or 4 days. | ||
| - | + | ===<p style="font-size:17px; background-color:#00dd77;">15.9 Seeding AAV293 for testing our Affibody and other VP-mVenus constructs</p>=== | |
| - | + | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;"> | + | |
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
| - | + | The pCerulean_VP1up_mVenus_Vp2/3 Construct will be tested in three different compositions.<br /> | |
<br /> | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| - | + | We want to test these constructs: | |
| + | <ul> | ||
| + | <li>pCerulean_VP1up_NLS_Affibody_VP2/3</li> | ||
| + | <li>pCerulean_VP1up_His_VP2/3</li> | ||
| + | <li>pCerulean_VP1up_mVenus_Vp2/3</li> | ||
| + | </ul> | ||
| + | The His-tag is for purify our constructs and in this case we want to test, if the particles are still infectious. Construct with the affibody will be tested for functionality and even for their better affinity to our EGFR overexpressing cell line A431. | ||
| + | |||
<br /> | <br /> | ||
<b>The results</b><br /> | <b>The results</b><br /> | ||
Interpretation and discussion of our results. | Interpretation and discussion of our results. | ||
<br /> | <br /> | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;">19.9 | + | |
| + | ===<p style="font-size:17px; background-color:#00dd77;">19.9 Seeding AAV293 for checking two different Rep/Cap Constructs</p>=== | ||
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
| - | + | During the project 22 silent nucleotide exchange mutations were introduced into the capsid coding construct to make it compatible with the RFC standards and to have single cutting restriction enzymes flanking the 453 and the 587 loop sequence. Two point mutations had to be dismissed, because either a first test transduction showed that the construct was not working anymore or the insertion of the synthesized gene posed serious problems, because the restriction enzyme did not work. | |
<br /> | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| - | + | Transfection and transduction was performed according to standard protocol. | |
| + | |||
<br /> | <br /> | ||
<b>The results</b><br /> | <b>The results</b><br /> | ||
| - | + | [[Image:Freiburg10 CC 19.9 FACS Checking functionality of REP CAP.jpg|800px]] | |
| + | <br /><br /> | ||
| + | Finally, we have a construct that is shown to produce infectious particles comparable to the current AAV systems and carries 20 point mutations. Now we can announce that the Adeno-associated Virus is compatible to the RFC standard and the idea to replace the loop sequences via ViralBricks works! | ||
<br /> | <br /> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 1,469: | Line 1,496: | ||
<b>The results</b><br /> | <b>The results</b><br /> | ||
As excepted, the expression of the CMV_mVenus and CMV_eGFP was very high even at 16h post transfection. The pTERT_mVenus construct worked aswell, but significantly weaker! | As excepted, the expression of the CMV_mVenus and CMV_eGFP was very high even at 16h post transfection. The pTERT_mVenus construct worked aswell, but significantly weaker! | ||
| - | Problematic is the fact, that there was no expression in HT1080 and A431 detectable. Due the fact that these cell lines are not optimized for CaCl<sub>2</sub> transfection. | + | Problematic is the fact, that there was no expression in HT1080 and A431 detectable. Due the fact that these cell lines are not optimized for CaCl<sub>2</sub> transfection. We can't evaluate if the bad transfection efficiency or the low promoter activity of pTERT are responsible for the results. |
| - | + | ||
<br /> | <br /> | ||
| Line 1,476: | Line 1,502: | ||
<br /> | <br /> | ||
<b>The motivation</b> | <b>The motivation</b> | ||
| - | <br /> Test the 18 constructs for functionality on A431 and HT1080 cells.<br /> | + | <br /> Test the following 18 constructs for functionality on A431 and HT1080 cells. Each of these constructs getting transfected in different compositions 25%, 50%, 75% and 90% with VP2_knockout 75%. In this way we are able to create viral particles with this ratios of VP proteins. We hope that the created particles are still infectious.<br /> |
| + | <ul> | ||
| + | <li>pCerulean_CFP_MiddleLinker_VP2/3insCap P528</li> | ||
| + | <li>pCerulean_Zegfr:1907_ShortlLinker_VP2/3insCap P527</li> | ||
| + | <li>pCerulean_6xHis_Middlelinker_VP2/3insCap P534</li> | ||
| + | <li>pCerulean_Zegfr:1907_Middlelinker_VP2/3insCap P535</li> | ||
| + | <li>pCerulean_Zegfr:1907_SEG_VP2/3_Capins 10:90 P502</li> | ||
| + | <li>pCerulean_Zegfr:1907_LongLinker_VP2/3_CapIns P503</li> | ||
| + | </ul> | ||
| + | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
<table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| Line 1,509: | Line 1,544: | ||
<br> [[Image:Freiburg 10 FACS Linker Libary.jpg|800px]] | <br> [[Image:Freiburg 10 FACS Linker Libary.jpg|800px]] | ||
<br /> | <br /> | ||
| + | Obviously the constructs still work! | ||
===<p style="font-size:17px; background-color:#00dd77;">22.9 Seeding cells for testing pTERT_mVenus in R/C P326 and P431 </p>=== | ===<p style="font-size:17px; background-color:#00dd77;">22.9 Seeding cells for testing pTERT_mVenus in R/C P326 and P431 </p>=== | ||
<br /> | <br /> | ||
| - | <b>The motivation</b> | + | <b>The motivation</b><br /> |
| - | We want to check if the pTERT-promoter is realiable for tumor specific gene expression. <br /><br /> | + | We want to check if the pTERT-promoter is realiable for tumor specific gene expression, we use two different kinds of R/C. <br /><br /> |
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
<table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| Line 1,549: | Line 1,585: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | <br /> | ||
| + | <b>The results</b> | ||
<br /> | <br /> | ||
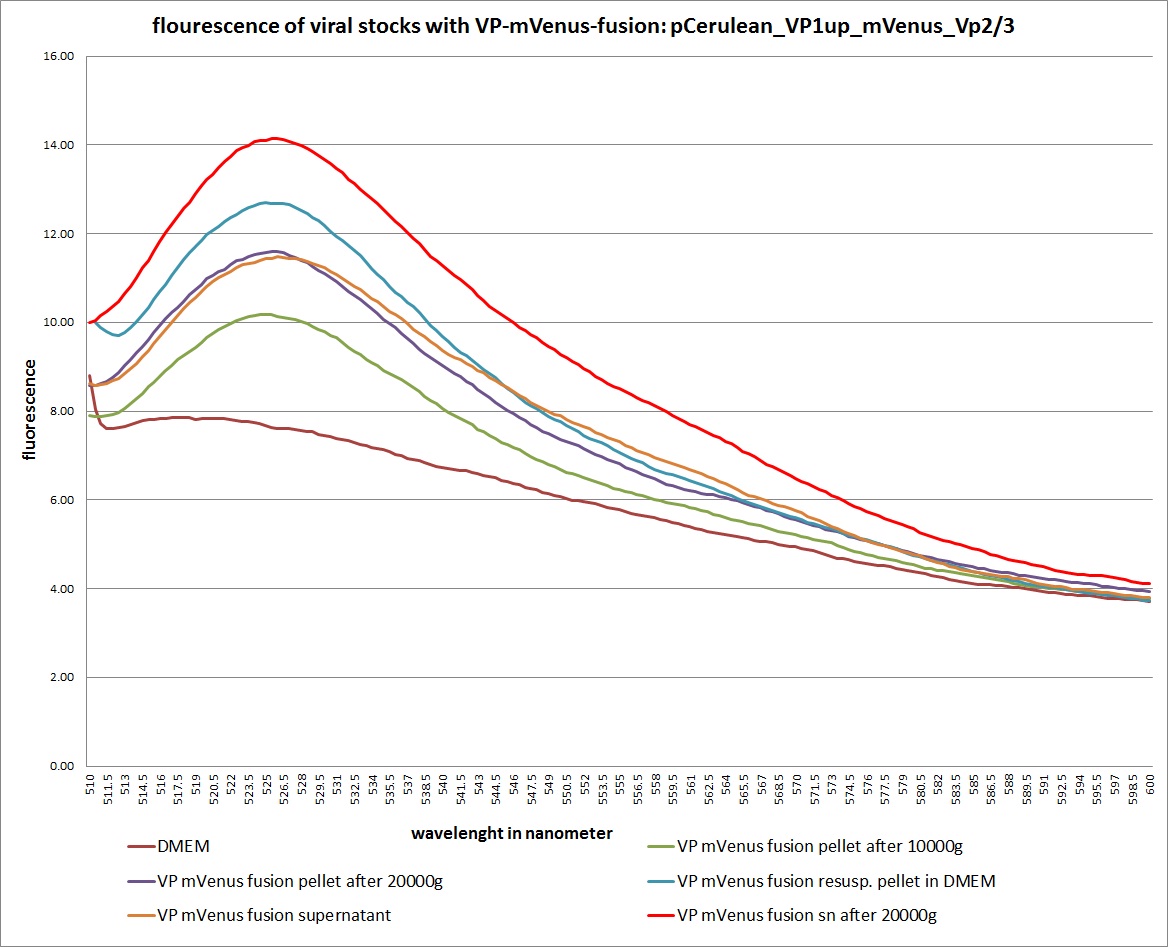
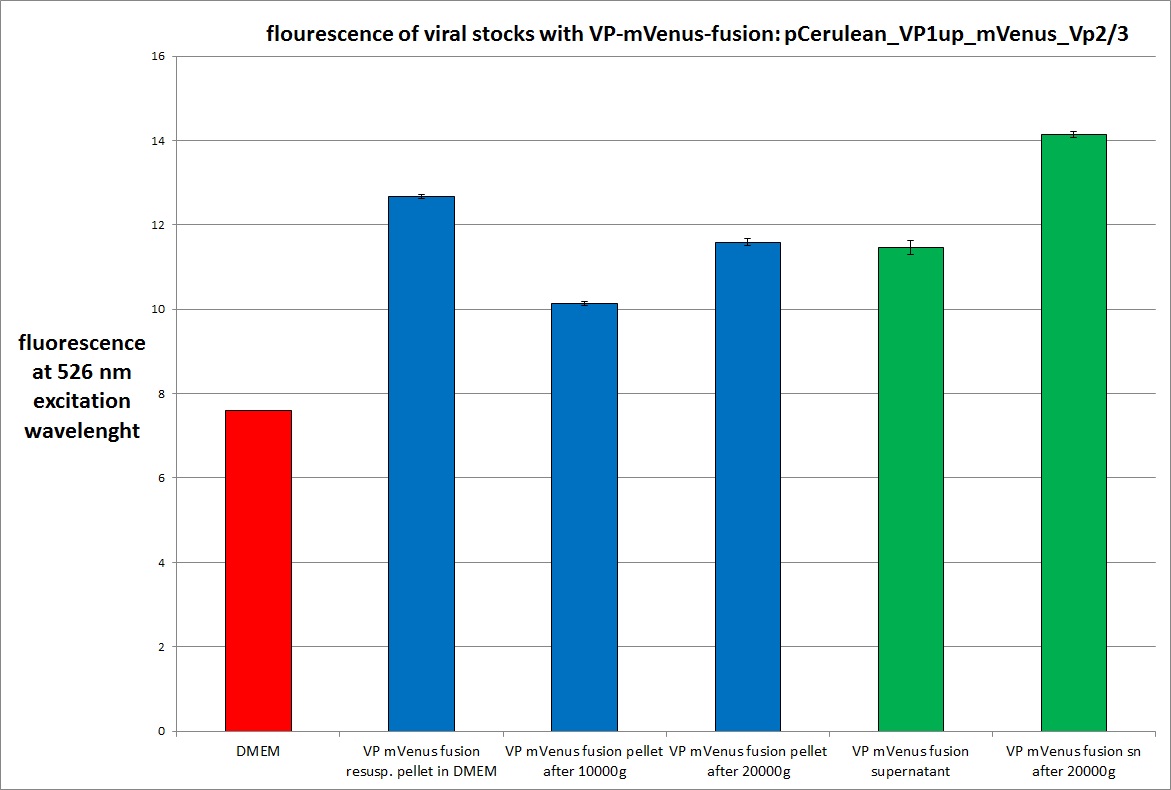
| + | ===<p style="font-size:17px; background-color:#00dd77;">29.9 Checking location of VP proteins via fluorescence microscopy and checking fractions of centrifugation steps via fluorescence spectrometer</p>=== | ||
<br /> | <br /> | ||
| - | <b>The | + | <b>The motivation</b>: We want to figure out if it is possible to purify our viral stocks via centrifugation with 10.000 g and/or 20.000 g. In theory/ according to the literature the viral particles should be in the cells and attached to the HSPGs at the cellular surfaces. |
<br /> | <br /> | ||
| + | <b>The plan</b>: Two different viral stocks were prepared: | ||
| + | <br /><br /> | ||
| + | Stock 1 with VP2-mVenus-fusion-Capsid loaded with TKGMK (4 wells from a 6 well plate)<br /> | ||
| + | <ul> | ||
| + | <li>R/C 1: 50% pCerulean_VP1up_NLS_mVenus_Vp2/3 P501</li> | ||
| + | <li>R/C 2: 50% Rep/Cap2: P449 pAAV_RC_4fachmut_VP1-ko:</li> | ||
| + | <li>GOI: TKGMK: P82.b</li> | ||
| + | <li>pHelper</li> | ||
| + | </ul> | ||
| + | <br /> | ||
| + | Stock 2 "standard" virus (4 wells from a 6 well plate)<br /> | ||
| + | <ul> | ||
| + | <li>R/C 1 P486</li> | ||
| + | <li>GOI: TKGMK: P82.b</li> | ||
| + | <li>pHelper</li> | ||
| + | </ul> | ||
| + | These two stocks got harvested according to standard protocol (scratching cells, transfer them in to 15 ml falcons, resuling 12ml solution total). After centrifugation (10 min 200g), the supernatant got transferred into two new 15 ml falcons and the pellets were resuspended with 10 ml DMEM Medium. The Stocks got freezed thawed two times so finally there were: | ||
| + | <ol> | ||
| + | <li>DMEM as negative control</li> | ||
| + | <li>resuspended Pellet with mVenus-fusioned viral capsids</li> | ||
| + | <li>resuspended Pellet with "standard virus"</li> | ||
| + | <li>supernatant with mVenus-fusioned viral capsids</li> | ||
| + | <li>supernatant with "standard virus"</li> | ||
| + | </ol> | ||
| + | <br /> | ||
| + | The fluorescence of each stock was measured via spectrometer. After that, stocks 2 and 4 were centrifugated 10min with 10.000 g followed by an other fluorescence measuring. Finally the stocks got spin down with 20.000 g for 10 min | ||
| + | <br /> | ||
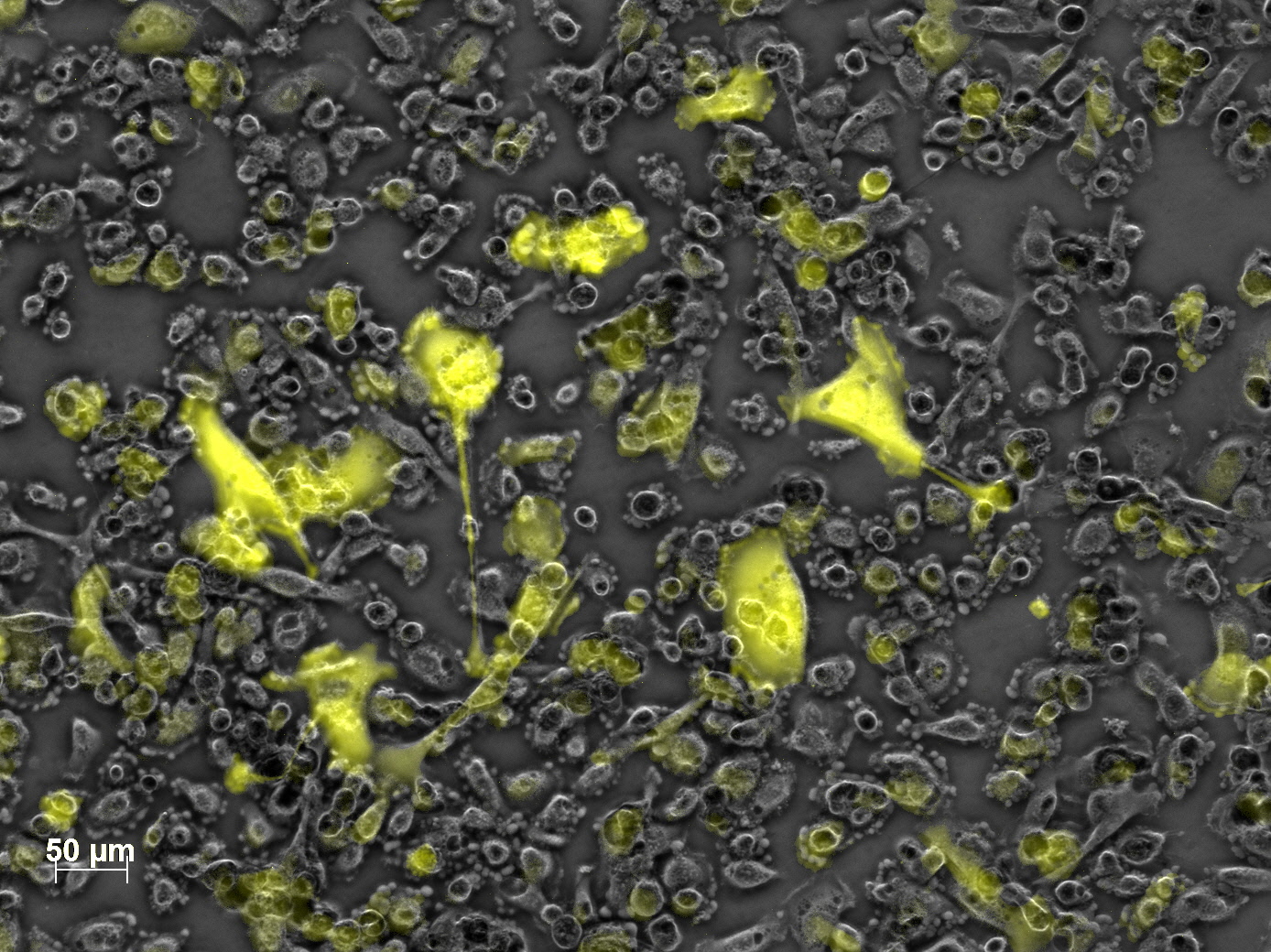
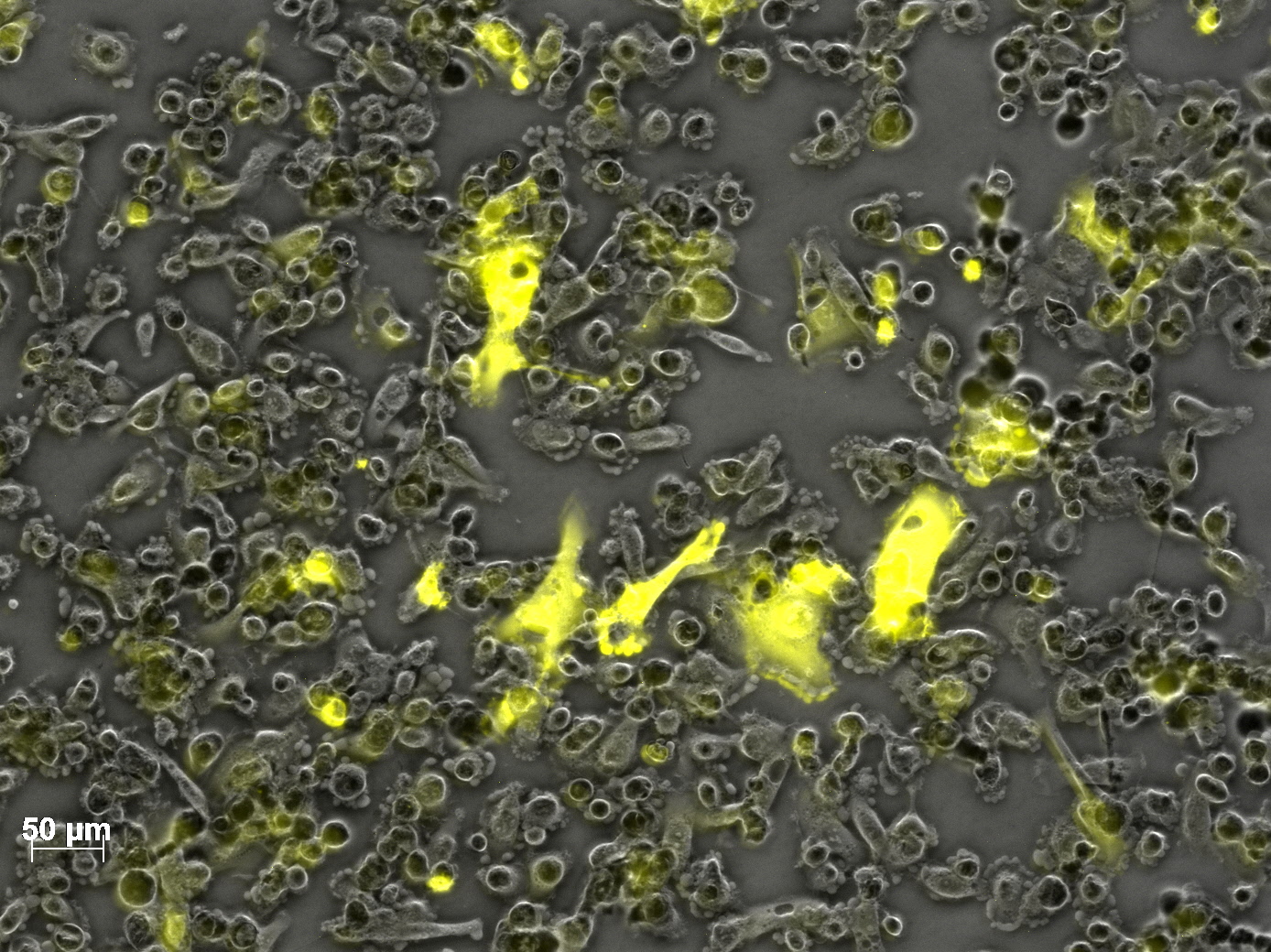
| + | <b>The results</b>: | ||
| + | <br /> | ||
| + | <gallery widths=300px heights=300px perrow=2 caption="Transfection with VP-mVenus-fusion constructs"> | ||
| + | Image:Freiburg10 VP mVenus fusion localisation 1.10 Close Picture2.jpg|Picture two days post transfection. | ||
| + | </gallery> | ||
| - | |||
<br /> | <br /> | ||
| - | + | [[Image: Freiburg10 mVenus VP fusion fluorescence centrifugation experiment graph.jpg|thumb|center|800px]] | |
<br /> | <br /> | ||
| - | <b>The | + | [[Image:Freiburg10 mVenus VP fusion fluorescence centrifugation experiment 526nm.jpg|thumb|center|800px]] |
| + | <br /><b>The conclusions</b>: | ||
| + | The highest amount of fluorescence was measured in the supernatant, the individual centrifugation steps had <b>no decrease in fluorescence</b> as consequence! | ||
| + | |||
<br /> | <br /> | ||
| + | Keep in mind, without further investigation it is not valid to say that we can purify our stocks via 10-20.000 g centrifugation steps, because we dont know if the viral capsids are still intact! The FACS analysis are not sufficient enough to make a decision, because our last samples had YFP as GOI (=> so efficient cell sorting was not possible). We need to do qPCRs to make a valid evidence. | ||
| + | <br /> | ||
| + | <br /> | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">30.9 Production of new Standard mVenus Vector</p>=== | ||
| + | <br /> | ||
| + | <b>The motivation</b><br /> | ||
| + | The AAV2 with 4 point mutations, inserted rep and cap and remutated KpnI restriction has to be checked for its functionality in order to compare it with the wild type AAV2. Hopefully there will be no significant difference.<br /> | ||
| + | <b>The plan</b><br /> | ||
| + | After the "delay" 200.000 A431 and HT1080 cells were seeded and transduced on the following day with 0,5 ml of this AAV2 mutant. | ||
<table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
<th width="100">Days</th> | <th width="100">Days</th> | ||
| - | |||
| - | |||
<th width="100">30.9</th> | <th width="100">30.9</th> | ||
<th width="100">1.10</th> | <th width="100">1.10</th> | ||
| Line 1,574: | Line 1,655: | ||
<th width="100">5.10</th> | <th width="100">5.10</th> | ||
<th width="100">6.10</th> | <th width="100">6.10</th> | ||
| + | <th width="100">7.10</th> | ||
| + | <th width="100">8.10</th> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,583: | Line 1,666: | ||
<td>-</td> | <td>-</td> | ||
<td>Seeding HT</td> | <td>Seeding HT</td> | ||
| - | <td>Harvest | + | <td>Harvest</td> |
| - | <td> | + | <td>delayed because of cell death</td> |
| - | <td> | + | <td>delayed because of cell death</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| - | <b>The results</b> | + | <br /> |
| - | + | <b>The results</b><br /> | |
| + | The stocks were harvested successfully | ||
| + | <br /> | ||
| - | + | ===<p style="font-size:17px; background-color:#00dd77;">30.9 Seeding AAV293 for production of standard TKGMK vector </p>=== | |
| - | + | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;"> | + | |
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
| - | + | We want to create a big amount of viral particles with TKGMK as gene of interest for using it as an standard vector for the MTT-assay. | |
<br /> | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| - | + | ||
| + | |||
| + | |||
<table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
<th width="100">Days</th> | <th width="100">Days</th> | ||
| - | |||
<th width="100">30.9</th> | <th width="100">30.9</th> | ||
<th width="100">1.10</th> | <th width="100">1.10</th> | ||
<th width="100">2.10</th> | <th width="100">2.10</th> | ||
<th width="100">3.10</th> | <th width="100">3.10</th> | ||
| + | <th width="100">4.10</th> | ||
| + | <th width="100">5.10</th> | ||
| + | <th width="100">6.10</th> | ||
| + | <th width="100">7.10</th> | ||
| + | <th width="100">8.10</th> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,614: | Line 1,703: | ||
<td>-</td> | <td>-</td> | ||
<td>Transfection</td> | <td>Transfection</td> | ||
| - | <td> | + | <td>-</td> |
| - | <td> | + | <td>-</td> |
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest</td> | ||
| + | <td>delayed because of cell death</td> | ||
| + | <td>delayed because of cell death</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
<br /> | <br /> | ||
<b>The results</b><br /> | <b>The results</b><br /> | ||
| - | + | The viral stocks were harvested efficiently and are ready to use. | |
<br /> | <br /> | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;">30.9 | + | ===<p style="font-size:17px; background-color:#00dd77;">30.9 Seeding AAV293 for production of vectors with mVenus missing beta globin</p>=== |
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
| - | + | Beta globin is said to improve (stratagene) the expression so we expect a worse mVenus (reportergene) expression compared to the standard AAVs. Therefore the seeded A431 and HT1080 cells were transduced with a AAV2 mutant missing beta-globin. | |
| + | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| - | + | Ater the "delay" 200.000 HT1080 and A431 cells were seeded and transduced with 0,5 ml AAV2 mutant on the following day. | |
<table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
| Line 1,660: | Line 1,754: | ||
<br /> | <br /> | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;">30.9 Seeding AAV293 for production of | + | ===<p style="font-size:17px; background-color:#00dd77;">30.9 Seeding AAV293 for production of vectors with mVenus missing HGH</p>=== |
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
| - | + | HGH is responsible for the mRNA polyadenylation of the virus so we expect expect a worse mVenus (reportergene) expression compared to the standard AAVs. Therefore the seeded A431 and HT1080 cells were transduced with a AAV2 mutant missing HGH. | |
<br /> | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| - | + | After the "delay" 200.000 A431 and HT1080 cells were seeded and transduced on the following day with 0,5 ml AAV2 mutant missing HGH. | |
<table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
| Line 1,698: | Line 1,792: | ||
<br /> | <br /> | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;">30.9 Seeding AAV293 for | + | ===<p style="font-size:17px; background-color:#00dd77;">30.9 Seeding AAV293 for testing RC with HSPG knockout for functionality</p>=== |
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
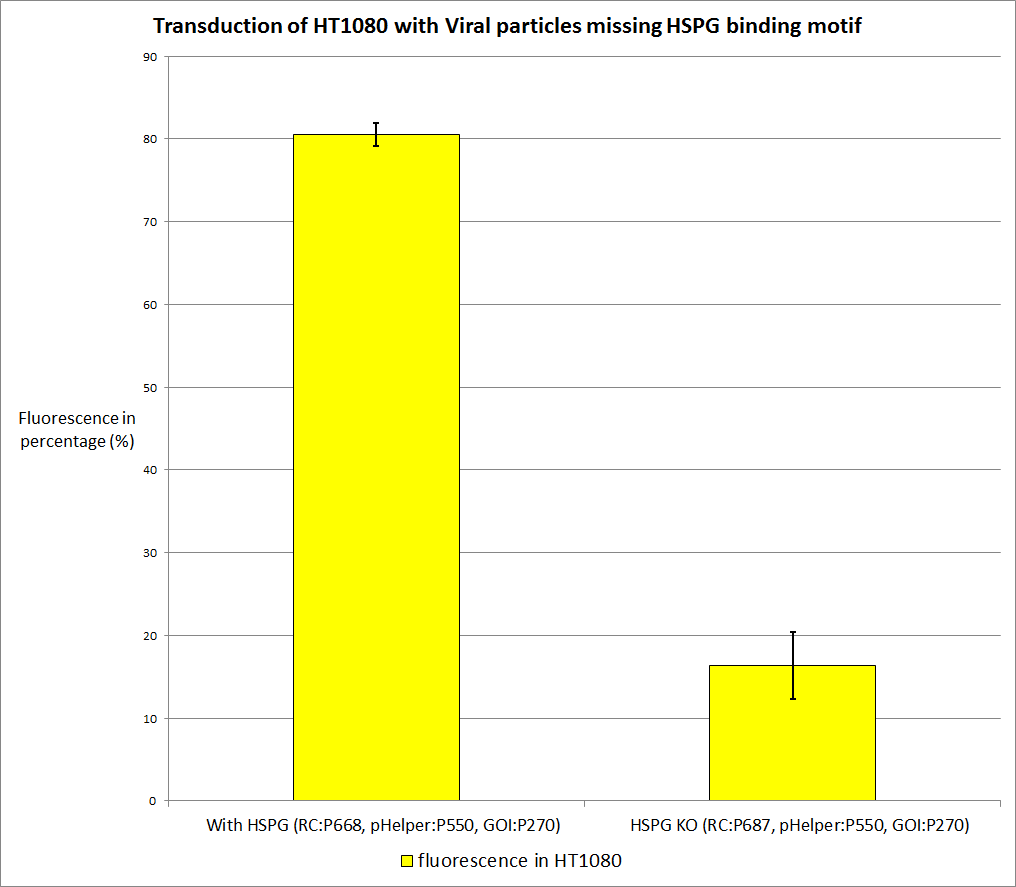
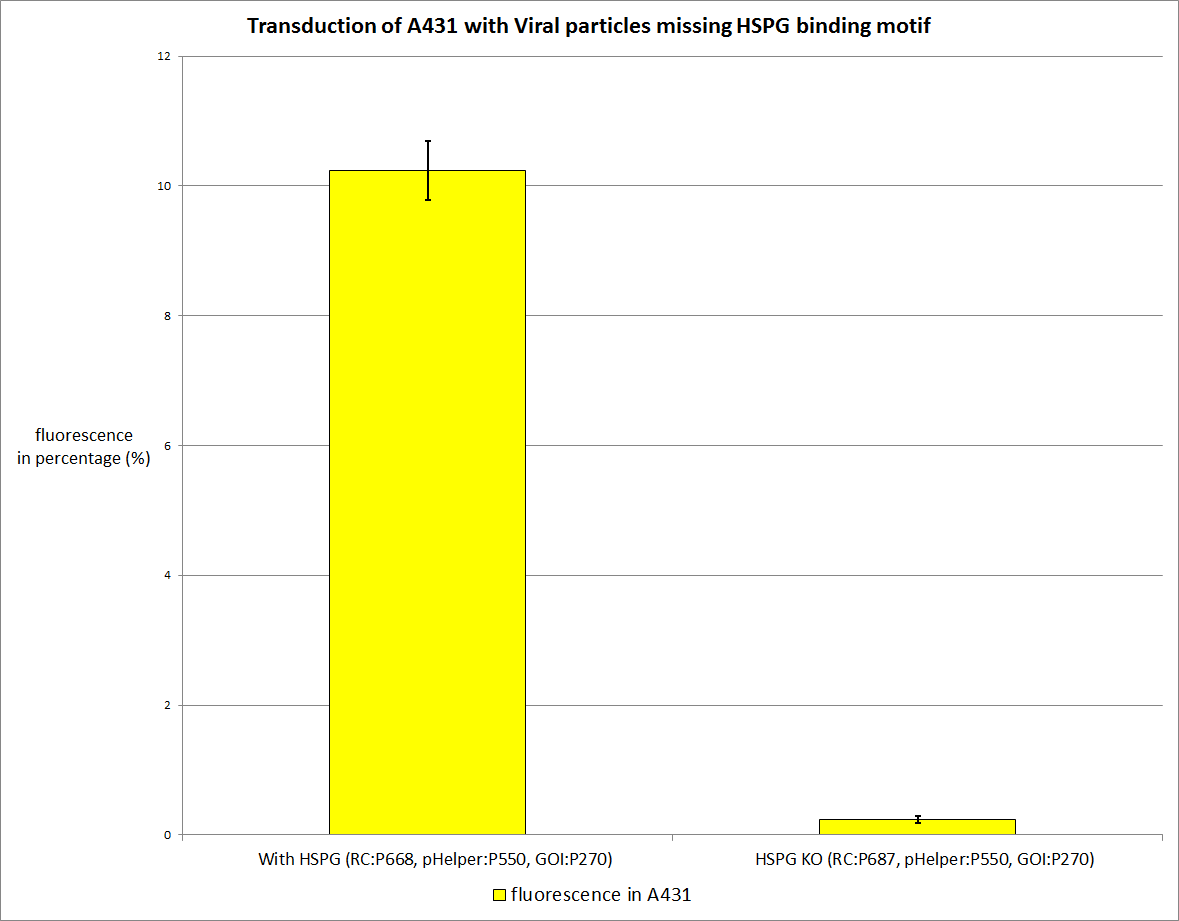
| - | + | The primary AAV2 receptor (the HSPG receptor) was knocked out so we expect a worse tranduction rate (about 0,3 to 0,4 of the standard transduction rate, assuming that there are HSPGs on the HT1080 and A431 cell surface). The worse the better. | |
<br /> | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| - | + | After the "delay" 200.000 A431 and HT1080 cells were seeded and transduced on the following day with 0,5 ml AAV2 mutant with a HSPG-receptor knockout. | |
<table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
| Line 1,733: | Line 1,827: | ||
<br /> | <br /> | ||
<b>The results</b><br /> | <b>The results</b><br /> | ||
| - | + | [[Image:Freiburg10 HT1080 missing HSPG binding motif.png|thumb|center|800px]] | |
| + | [[Image:Freiburg10 A431 missing HSPG binding motif.png|thumb|center|800px]] | ||
<br /> | <br /> | ||
| + | There is a significant difference in transduction efficiency. The actual reduction is even higher than expected, according to literature it should be around 60%, we reached 80%. We measured via qPCR the amount of viral particles for each stock, and they are nearly the same the HSPG Knockout has 2*10<sup>7</sup>particles per ml , the stock with HSPG-binding motif has 4.5*10<sup>6</sup> particles per ml. | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;">30.9 Seeding AAV293 for | + | ===<p style="font-size:17px; background-color:#00dd77;">30.9 Seeding AAV293 for testing RC WITHOUT HSPG knockout for functionality</p>=== |
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
| - | + | The produced vector has the intact HSPG-binding motif, so we can compare it with the vector produced in: "Seeding AAV293 for testing RC with HSPG knockout for functionality" | |
<br /> | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| - | + | Transfection and transduction were performed according to standard protocol. | |
<table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
| Line 1,771: | Line 1,867: | ||
<br /> | <br /> | ||
<b>The results</b><br /> | <b>The results</b><br /> | ||
| - | + | See 30.9 Seeding AAV293 for testing RC with HSPG knockout for functionality | |
<br /> | <br /> | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;"> | + | ===<p style="font-size:17px; background-color:#00dd77;">12.10 Seeding AAV293 cells for transfection with VP-BAP</p>=== |
<br /> | <br /> | ||
| - | <b>The motivation</b> | + | <b>The motivation</b> |
| - | + | ||
<br /> | <br /> | ||
| + | We want to create a viral stock for purification and biotinylation, the ratio of the Rep/Cap plasmids is 1:1.<br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| - | + | Transfection was performed according to standard protocol: | |
| + | <ul> | ||
| + | <li>Gene of interest: mVenus</li> | ||
| + | <li>R/C: pSB1C3-001_pCMV_[AAV]VP123_587_BAP in cotransfection with pSB1C3_001_RC_IRCK_P5tataless clone 1</li> | ||
| + | </ul> | ||
| + | |||
<table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
<th width="100">Days</th> | <th width="100">Days</th> | ||
| - | <th width="100"> | + | <th width="100">12.10</th> |
| - | <th width="100"> | + | <th width="100">13.10</th> |
| - | <th width="100"> | + | <th width="100">14.10</th> |
| - | <th width="100"> | + | <th width="100">15.10</th> |
| - | <th width="100"> | + | <th width="100">16.10</th> |
| - | <th width="100"> | + | <th width="100">17.10</th> |
| - | <th width="100"> | + | <th width="100">18.10</th> |
| - | <th width="100"> | + | <th width="100">19.10</th> |
| - | <th width="100"> | + | <th width="100">20.10</th> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,802: | Line 1,903: | ||
<td>-</td> | <td>-</td> | ||
<td>Seeding HT</td> | <td>Seeding HT</td> | ||
| - | <td>Harvest</td> | + | <td>Harvest + Transduction</td> |
| - | <td> | + | <td>-</td> |
| - | <td> | + | <td>FACS</td> |
</tr> | </tr> | ||
</table> | </table> | ||
| + | <b>The results</b> | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">12.10 Seeding AAV293 cells for transfection with mVenus as gene of interest and pCerulean_VP1up_NLS_mVenus_VP2/3_HSPG-KO</p>=== | ||
<br /> | <br /> | ||
| - | <b>The | + | <b>The motivation</b> |
| - | + | ||
<br /> | <br /> | ||
| + | We want to create a viral stock for purification and biotinylation, the ratio of the Rep/Cap plasmids is 1:1.<br /> | ||
| + | <b>The plan</b><br /> | ||
| + | Transfection was performed according to standard protocol: | ||
| + | <ul> | ||
| + | <li>Gene of interest: mVenus</li> | ||
| + | <li>R/C: pCerulean_VP1up_NLS_mVenus_VP2/3_HSPG-KO in cotransfection with pSB1C3_001_RC_IRCK_VP1-ko_HSPG-ko_P5tataless cl1</li> | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;"> | + | </ul> |
| + | |||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">12.10</th> | ||
| + | <th width="100">13.10</th> | ||
| + | <th width="100">14.10</th> | ||
| + | <th width="100">15.10</th> | ||
| + | <th width="100">16.10</th> | ||
| + | <th width="100">17.10</th> | ||
| + | <th width="100">18.10</th> | ||
| + | <th width="100">19.10</th> | ||
| + | <th width="100">20.10</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>-</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest + Transduction</td> | ||
| + | <td>-</td> | ||
| + | <td>FACS</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <b>The results</b> | ||
| + | |||
| + | |||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">12.10 Seeding AAV293 cells for transfection with mVenus as GOI and CFP_Middlelinker_VP2/3_HSPG-KO</p>=== | ||
<br /> | <br /> | ||
<b>The motivation</b><br /> | <b>The motivation</b><br /> | ||
| - | This | + | This viral stock is for live imaging and probably for western blotting, the ratio of the Rep/Cap plasmids is 1:1.<br /> |
| + | <b>The plan</b><br /> | ||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">18.10</th> | ||
| + | <th width="100">19.10</th> | ||
| + | <th width="100">20.10</th> | ||
| + | <th width="100">21.10</th> | ||
| + | <th width="100">22.10</th> | ||
| + | <th width="100">23.10</th> | ||
| + | <th width="100">24.10</th> | ||
| + | <th width="100">25.10</th> | ||
| + | <th width="100">26.10</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>-</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest + Transduction</td> | ||
| + | <td>-</td> | ||
| + | <td>FACS</td> | ||
| + | </tr> | ||
| + | </table><br /> | ||
| + | Transfection was performed according to standard protocol: | ||
| + | <ul> | ||
| + | <li>Gene of interest: mVenus</li> | ||
| + | <li>R/C: CFP_Middlelinker_VP2/3_HSPG-KO in cotransfection with pSB1C3_001_RC_IRCK_P5tataless clone 1</li> | ||
| + | <br /> | ||
| + | </ul> | ||
| + | <b>The results</b> | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">17.10 Production of AAV2 with 587-His</p>=== | ||
| + | Investigator Patrick <br> | ||
| + | <br> | ||
| + | <b>The motivation</b><br /> | ||
| + | We wanted to produce AAV2 which could be purified via affinity purification. Therefore 10 x 10cm^2 were seeded with AAV-293. Five of these plates contained serum-free medium and cells accustomized to this. <br> | ||
| + | Used plasmids: | ||
| + | * pHelper | ||
| + | * pSB1C3_leftITR_CMV_betaglobin_mvenus_hGH_rightITR (P541) | ||
| + | * 50% pSB1C3_001_RC_IRCK_P5tataless (P712) | ||
| + | * 50% pSB1C3-001_pCMV_[AAV2]VP123_587_HIS (P638) | ||
| + | |||
<br /> | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
In this section, the design of the experiment is explained. | In this section, the design of the experiment is explained. | ||
| + | <br /> | ||
<table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
<tr> | <tr> | ||
<th width="100">Days</th> | <th width="100">Days</th> | ||
| - | <th width="100"> | + | <th width="100">17.10</th> |
| - | <th width="100"> | + | <th width="100">19.10</th> |
| - | <th width="100"> | + | <th width="100">20.10</th> |
| - | <th width="100"> | + | <th width="100">21.10</th> |
| - | <th width="100"> | + | <th width="100">22.10</th> |
| - | <th width="100">5.10</th> | + | <th width="100">23.10</th> |
| - | <th width="100"> | + | <th width="100">24.10</th> |
| - | <th width="100"> | + | <th width="100">25.10</th> |
| - | <th width="100"> | + | <th width="100">26.10</th> |
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>AAV2 harvest</td> | ||
| + | <td>Transduction</td> | ||
| + | <td></td> | ||
| + | <td>FACS</td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | </table><br /> | ||
| + | |||
| + | <b>The results</b><br /> | ||
| + | Interpretation and discussion of our results. | ||
| + | <br /> | ||
| + | |||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">18.10 Seeding AAV293 cells for transfection Superconstructs</p>=== | ||
| + | <br /> | ||
| + | <b>The motivation</b><br /> | ||
| + | We want to create a viral stock for purification and biotinylation, the ratio of the Rep/Cap plasmids is 1:1.<br /> | ||
| + | <b>The plan</b><br /> | ||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">18.10</th> | ||
| + | <th width="100">19.10</th> | ||
| + | <th width="100">20.10</th> | ||
| + | <th width="100">21.10</th> | ||
| + | <th width="100">22.10</th> | ||
| + | <th width="100">23.10</th> | ||
| + | <th width="100">24.10</th> | ||
| + | <th width="100">25.10</th> | ||
| + | <th width="100">26.10</th> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,840: | Line 2,065: | ||
<td>-</td> | <td>-</td> | ||
<td>Seeding HT</td> | <td>Seeding HT</td> | ||
| - | <td>Harvest</td> | + | <td>Harvest + Transduction</td> |
| - | <td> | + | <td>-</td> |
| - | <td> | + | <td>FACS</td> |
| + | </tr> | ||
| + | </table><br /> | ||
| + | Transfection was performed according to standard protocol:<br /> | ||
| + | <br /> | ||
| + | <b>Gene of interest: CMV_mVenus</b> | ||
| + | <ul> | ||
| + | <li>pSB1C3_CMV_Zegfr:1907_MiddleLinker_VP2/3_587-KO_6xHis in ratio 1:1 pSB1C3_001_RC_IRCK_VP2-ko_HSPG-ko_P5tataless cl1</li> | ||
| + | <li>pSB1C3-001_pCMV_[AAV2]VP123_453_Z34C in ratio 1:1 pSB1C3_001_RC_IRCK_HSPG-ko_P5tataless_RFC10</li> | ||
| + | <li>pSB1C3_001_CMV_VP123_587-KO_Z34C in ratio 1:1 pSB1C3_001_RC_IRCK_HSPG-ko_P5tataless_RFC10</li> | ||
| + | <li> pSB1C3_001_CMV_VP123_587-KO_Z34C_spacer in ratio 1:1 pSB1C3_001_RC_IRCK_HSPG-ko_P5tataless_RFC10</li> | ||
| + | </ul><br /> | ||
| + | <b>Gene of interest: pTERT_TKGMK</b> | ||
| + | <ul> | ||
| + | <li>pSB1C3_CMV_Zegfr:1907_MiddleLinker_VP2/3_587-KO_6xHis in ratio 1:1 pSB1C3_001_RC_IRCK_VP2-ko_HSPG-ko_P5tataless cl1</li> | ||
| + | </ul><br /> | ||
| + | <b>Gene of interest: CMV_TKGMK</b> | ||
| + | <ul> | ||
| + | <li>pSB1C3_CMV_Zegfr:1907_MiddleLinker_VP2/3_587-KO_6xHis in ratio 1:1 pSB1C3_001_RC_IRCK_VP2-ko_HSPG-ko_P5tataless cl1</li> | ||
| + | </ul><br /> | ||
| + | |||
| + | <b>The results</b> | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">18.10 Seeding AAV293 cells for transfection with Cytosindeaminase</p>=== | ||
| + | <br /> | ||
| + | <b>The motivation</b><br /> | ||
| + | <br /> | ||
| + | We want to create viral stocks with CMV_Cytosindeaminase and pTERT_Cytosindeaminase as gene of interests, to check qualitatively the bystander effect.<br /> | ||
| + | <br /><b>The plan</b><br /> | ||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">18.10</th> | ||
| + | <th width="100">19.10</th> | ||
| + | <th width="100">20.10</th> | ||
| + | <th width="100">21.10</th> | ||
| + | <th width="100">22.10</th> | ||
| + | <th width="100">23.10</th> | ||
| + | <th width="100">24.10</th> | ||
| + | <th width="100">25.10</th> | ||
| + | <th width="100">26.10</th> | ||
| + | <th width="100">27.10</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>seeding AAV293</td> | ||
| + | <td>-</td> | ||
| + | <td>-</td> | ||
| + | <td>Transfection</td> | ||
| + | <td>-</td> | ||
| + | <td>-</td> | ||
| + | <td>Seeding HT</td> | ||
| + | <td>Harvest + Transduction</td> | ||
| + | <td>-</td> | ||
| + | <td>FACS</td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | Transfection, harvest and transduction were performed according to standard protocol:<br /> | ||
<br /> | <br /> | ||
| - | <b> | + | <b>Gene of interest: CMV_Cytosindeaminase</b> |
| - | + | <ul> | |
| + | <li>P712</li> | ||
| + | </ul> | ||
| + | <b>Gene of interest: pTERT_Cytosindeaminase</b> | ||
| + | <ul> | ||
| + | <li>P712</li> | ||
| + | </ul> | ||
<br /> | <br /> | ||
| - | ===<p style="font-size:17px; background-color:#00dd77;"> | + | <b>The results</b> |
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">22.10.2010 Transduction with old mVenus Stocks for taking pictures</p>=== | ||
<br /> | <br /> | ||
<b>The motivation</b> | <b>The motivation</b> | ||
| + | We transduced HT1080 with our stocks packaged with CMV_mVenus. | ||
<br /> | <br /> | ||
<b>The plan</b><br /> | <b>The plan</b><br /> | ||
| Line 1,862: | Line 2,151: | ||
<th width="100">24.9</th> | <th width="100">24.9</th> | ||
<th width="100">25.9</th> | <th width="100">25.9</th> | ||
| - | <th width="100">26. | + | </tr> |
| - | <th width="100">27. | + | <tr> |
| - | <th width="100">28. | + | <td>Action</td> |
| - | <th width=" | + | <td>seeding HT1080</td> |
| - | <th width="100"> | + | <td>Transduction</td> |
| + | <td>-</td> | ||
| + | <td>Fluorescence microscopy</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <b>The results</b> | ||
| + | |||
| + | <gallery widths=300px heights=300px perrow=2 caption="Transduction with vectors pSB1C3_001_RC_IRCK_P5tataless as R/C and CMV_mVenus"> | ||
| + | Image:Freiburg10_transduction_CMV_mVenus_1.png| | ||
| + | Image:Freiburg10_transduction_CMV_mVenus_2.png| | ||
| + | Image:Freiburg10_transduction_CMV_mVenus_4.png| | ||
| + | </gallery> | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">23.10 Testing the bystander effect of the cytosine deaminase</p>=== | ||
| + | <br /> | ||
| + | <b>The motivation</b><br /> | ||
| + | <br /> | ||
| + | The activated form of 5-fluorocytosine has the ability to diffuse through the plasma membrane. In this way cells get killed which were not transduced by the viral particles. We use an old yfp stock t determine if the transgene expression is on and to harvest the cells which are transduced with cytosine deaminase loaded AAV2.<br /> | ||
| + | <br /><b>The plan</b><br /> | ||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">23.10</th> | ||
| + | <th width="100">24.10</th> | ||
| + | <th width="100">25.10</th> | ||
| + | <th width="100">26.10</th> | ||
| + | <th width="100">27.10</th> | ||
| + | <th width="100">28.10</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>Transduce HT1080 and seed HT1080</td> | ||
| + | <td>Transfer the transduced HT1080 onto the non transduced HT1080</td> | ||
| + | <td>-</td> | ||
| + | <td>-</td> | ||
| + | <td>-</td> | ||
| + | <td>Measure living cells via Neubauer cellchamber</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <b>Gene of interest: CMV_Cytosindeaminase</b> | ||
| + | <ul> | ||
| + | <li>P712</li> | ||
| + | </ul> | ||
| + | <b>Gene of interest: CMV_mVenus</b> | ||
| + | <ul> | ||
| + | <li>P468</li> | ||
| + | </ul> | ||
| + | <br />All dishes had this order:<br /> | ||
| + | <br /> | ||
| + | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="80"> </th> | ||
| + | <th width="80">1</th> | ||
| + | <th width="80">2</th> | ||
| + | <th width="80">3</th> | ||
| + | |||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>A</td> | ||
| + | <td>Medium with prodrug</td> | ||
| + | <td>Medium with prodrug</td> | ||
| + | <td>Medium with prodrug</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>B</td> | ||
| + | <td>Medium</td> | ||
| + | <td>Medium</td> | ||
| + | <td>Medium</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br />The concentration of the prodrug 5-fluorocytosine is 54 mmolar<br /> | ||
| + | <br />Cells were seeded following this scheme:<br /> | ||
| + | <b><font size="3">plate 1</font></b> | ||
| + | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="80"> </th> | ||
| + | <th width="80">1</th> | ||
| + | <th width="80">2</th> | ||
| + | <th width="80">3</th> | ||
| + | |||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>A</td> | ||
| + | <td>1*10<sup>5</sup> HT1080 transduced with CMV_Cytosindeaminase</td> | ||
| + | <td>1*10<sup>5</sup> HT1080 transduced with CMV_Cytosindeaminase</td> | ||
| + | <td>1*10<sup>5</sup> HT1080 transduced with CMV_Cytosindeaminase</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>B</td> | ||
| + | <td>1*10<sup>5</sup> HT1080 transduced with CMV_Cytosindeaminase</td> | ||
| + | <td>1*10<sup>5</sup> HT1080 transduced with CMV_Cytosindeaminase</td> | ||
| + | <td>1*10<sup>5</sup> HT1080 transduced with CMV_Cytosindeaminase</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br /> | ||
| + | |||
| + | <br /> | ||
| + | <b><font size="3">plate 2</font></b> | ||
| + | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="80"> </th> | ||
| + | <th width="80">1</th> | ||
| + | <th width="80">2</th> | ||
| + | <th width="80">3</th> | ||
| + | |||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>A</td> | ||
| + | <td>2*10<sup>5</sup> HT1080 non transduced</td> | ||
| + | <td>2*10<sup>5</sup> HT1080 non transduced</td> | ||
| + | <td>2*10<sup>5</sup> HT1080 non transduced</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>B</td> | ||
| + | <td>2*10<sup>5</sup> HT1080 non transduced</td> | ||
| + | <td>2*10<sup>5</sup> HT1080 non transduced</td> | ||
| + | <td>2*10<sup>5</sup> HT1080 non transduced</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br /> | ||
| + | The next plates were seeded with non transduced cells and 1*10<sup>5</sup> of the cytosin deaminase transduced cells | ||
| + | <br /> | ||
| + | <b><font size="3">plate 3</font></b> | ||
| + | <table border="3" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="80"> </th> | ||
| + | <th width="80">1</th> | ||
| + | <th width="80">2</th> | ||
| + | <th width="80">3</th> | ||
| + | |||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>A</td> | ||
| + | <td>2*10<sup>5</sup> HT1080</td> | ||
| + | <td>2*10<sup>5</sup> HT1080</td> | ||
| + | <td>2*10<sup>5</sup> HT1080</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>B</td> | ||
| + | <td>2*10<sup>5</sup> HT1080</td> | ||
| + | <td>2*10<sup>5</sup> HT1080</td> | ||
| + | <td>2*10<sup>5</sup> HT1080</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | <b>The results</b> | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">25.10.2010 Transduction with several constructs for Flow Cytometry</p>=== | ||
| + | <br /> | ||
| + | <b>The motivation</b> | ||
| + | We want to quantify via flow cytometry the Affibody superconstruct, Darpin HSPG-KO Construct and standard R/C with HSPG-KO. | ||
| + | <br /> | ||
| + | <b>The plan</b><br /> | ||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">25</th> | ||
| + | <th width="100">26</th> | ||
| + | <th width="100">27</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Action</td> | ||
| + | <td>Transduction</td> | ||
| + | <td>-</td> | ||
| + | <td>FACS</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <b>The results</b> | ||
| + | |||
| + | ===<p style="font-size:17px; background-color:#00dd77;">BLANK EXPERIMENT</p>=== | ||
| + | <br /> | ||
| + | <b>The motivation</b> | ||
| + | <br /> | ||
| + | <b>The plan</b><br /> | ||
| + | <table border="5" rules="all" cellpadding="5" cellspacing="1" style="font-size:13pt;text-align:center" > | ||
| + | <tr> | ||
| + | <th width="100">Days</th> | ||
| + | <th width="100">-</th> | ||
| + | <th width="100">-</th> | ||
| + | <th width="100">-</th> | ||
| + | <th width="100">-</th> | ||
| + | <th width="100">-</th> | ||
| + | <th width="100">-</th> | ||
| + | <th width="100">-</th> | ||
| + | <th width="100">-</th> | ||
| + | <th width="100">-</th> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| Line 1,882: | Line 2,358: | ||
</table> | </table> | ||
<b>The results</b> | <b>The results</b> | ||
| + | |||
<h1>Citations</h1> | <h1>Citations</h1> | ||
Latest revision as of 21:38, 27 October 2010
- March (labday 1)
- April (labday 2 - 5)
- May (labday 6 - 17)
- June (labday 18 - 45)
- July (labday 46 - 75)
- August part 1 (labday 76 - 92)
- August part 2 (labday 93 - 106)
- September part 1 (labday 107 - 123)
- September part 2 (labday 124 - 135)
- October part 1 (labday 136 - 149 )
- October part 2 (labday 150 - 166 )
- November (labday 167 - 170 )
- Cellculture
Cellculture
This subpage is for cellculture, the structure of each experiment is following:
Date of first action, title of the experiment
The motivation
This short introduction provides an inside into our thoughts for this experiment.
The plan
In this section, the design of the experiment is explained.
The results
Interpretation and discussion of our results.
14.6 Seeding AAV293 for transfection
The motivation
We want to create viral stocks with the stratagene R/C, the gene of interest is mVenus. This is a qualitative control.
The plan
| Days | 14.6 | 15.6 | 16.6 | 17.6 | 18.6 | 19.6 | 20.6 | 21.6 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | Fluorescence microscopy |
AAV 293 and HT1080 Cells have been splitted and plated out on 10 cm dishes for transfektion. After harvesting the cells according to the standard protocol the cell-pellets were resuspendiated in 15ml of DYT medium. 2,5µl of this cell suspension have been mixed with 47.5µl of trypan blue and the cells were counted by using the Neubauer-Meteringchamber.
We counted 12,5x 10^6 cells/ml for the AAV293 cell line and 10x10^6 cells /ml for the HT1080 cell line.
1ml and 1,5 ml of the AAV 293 cells have been seeded on four dishes with 250µl of the cell suspension. Note that that the date is wrong on the plates and the flasks! Cells have been already plated out on the 14th. of June.
We also seated two flasks one for each cell line. Therefor we used 1ml of the HT1080 cell suspension and 2ml of the the AAV293 cells.
1)
- pAAV_iGEM_mVenus_YFP (glycerol stock B34, P39) P39 has the correct sequence (confirmed)
- pHelper
- pAAV_RC
2)
- pAAV_iGEM_mVenus_YFP (glycerol stock B33, P38) P38 (we dont know if P39 has the correct sequence!)was used because the amount of P39 was not sufficient enough for four Transfections!
- pHelper
- pAAV_RC
Plasmid concentrations:
pAAV_RC: 1 µg/µl
pHelper: 280 ng/µl
pAAV_iGEM_mVenus_YFP, P39: 180,83 ng/µl
pAAV_iGEM_mVenus_YFP, P38: 179,75 ng/µl
Cellculture clone 1 and 2 were transfected with P39. Cellculture clone 3 and 4 were transfected with P38.
We could only pipet 3,3 µg (instead of 10 µg) from each of the 3 plasmids into the 15 ml falcons due to insufficient amount of plasmid.
Adrian tried to examine the precipation of the CaCl2+ DNA Clusters. We have to optimize the pH of our 2xHBS at the moment it is 11.12.
The viral stocks were harvested according to standard protocol.
The results
According to the stratagene protocol, transduction efficencies about ~80% are achievable. We performed every step under guidance, so probably something is wrong with the buffers, cells or even the standard protocol. The next step is to repeat this transfection.
19.6 Seeding AAV293 for second transfection according to standard protocol
The motivation
We want to repeat the last transfection and obtain a vsibly higher GOI (mVenus) expression.
The plan
| Days | 19.6 | 20.6 | 20.6 | 21.6 | 22.6 | 23.6 | 24.6 | 25.6 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | Fluorescence microscopy |
Six transfections were performed according to the standart protocol:
Three 10 cm cellcluture dishes with 293 cells were transfected with 10 µg DNA (3,33 µg each plasmid) and the other three 10 cm cellculture dishes with 30 µg DNA (10 µg DNA each plasmid).
Plasmids:
- pAAV_iGEM-MCS_mVenus (P41), 507 ng/µl
- pAAV_RC , 1000 ng/µl
- pHelper , 280 ng/µl
- Transduction of 2x6 wells was successfully done (14.25)
- one six well was transduced with 10µg the other with 30µg DNA amount
The results
The Transduction efficency is still quite low! We have to optimize the stratagene protocol!
30.6 Examination of different Transduction methods
The motivation
We want to check whether the amount of our produced AAV2s has an effect on the transduction rate.
Instead of 500µl, 1000µl of the AAV-stock were pipetted into each well.
The plan
| Days | 30.6 | 1.7 | 2.7 | 3.7 | 4.7 | 5.7 | 6.7 | 7.7 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | Fluorescence microscopy |
1. Plate I(A is up)
| 3x10^5 cells | 3x10^5 cells | 3x10^5 cells |
| 3x10^5 cells | 3x10^5 cells | 3x10^5 cells |
Transduction:
| 1000 µl of Viral Stock DROPPED | 1000 µl of Viral Stock gently resuspended | 1500 µl of Viral Stock rough resuspending |
| 1000 µl of Viral Stock DROPPED | 1000 µl of Viral Stock gently resuspended | no Transduction |
The results
All cells died probably because we forgot to wash the cells with PBS before adding fresh medium. It was not possible to take pictures because the dead cells were all clumped.
Looks like we should not transduce 3x10^5 cells with 1000 µl (or more) of our viral solution because almost all cells died in all approaches and the medium indicator was yellow.
So we can not say if resuspendig the cells with the viral soultion is a useful alteration of the standard protocol.
3.7 Seeding AAV293 for transfection with different plasmid amounts and harvesting methods
The motivation
We want to check if the amount of plasmids is the reason for the low transduction efficiency. Unfortunately the flow cytometry (FACS) is not available at the moment so the results can only be checked via fuorescence microscopy.
The plan
| Days | 3.7 | 4.7 | 5.7 | 6.7 | 7.7 | 8.7 | 9.7 | 10.7 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | Fluorescence microscopy |
Transfection with P41 (pAAV_iGEM-MCS_mVenus) was performed according standart protocol.
Two transfections where carried out with 10µg (3,33 µg each plasmid) DNA and the other two with 20µg DNA (6,66 µg each plasmid).
We want to investigate which of the following approaches yields a better transduction efficiency.
- Half of the transfected cells were exposed 3 cycles of thawing and freezing and then centrifuged (15 ml falcon, 2100G). The supernatant was transfered into a new 15 ml falcon and used for transduction.
- The other half was centrifuged at 300 G for 5min . The supernatant was transfered into another 15 ml falcon and the pellet was resuspended with 5 ml of DMEM. The content of these two falcons was also exposed to 3 cycles of thawing and freezing and then used for transduction.
- We have got 2 10 cm cellculture dishes with 10µg and 2 dishes with 20µg DNA used for transfection.
Approach with standart protocol (one dish with 10 and the other with 20µg Plasmids)(2100 G and 3 cycles of freezing and thawing)
Deviations from the standart protocol:
- The cells were centrifuged at 2100 G instead of 10.000 G
- The viruses were harvested after 44 hours
- 3 cycles of freezing and thawing
- used plasmids: 10µg, 20µg GOI-plasmid (mVenus)
- we have 10µg and 20µg from standart protocol => standard virus
- we have 10µg and 20µg from standart protocol pellet => Pellet
- we have 10µg and 20µg from standart protocol suspension => Super
- amount of 500µl
1. Plate I(A is up)
| 10µg dropped Super | 10µg dropped Super gently resuspending | 10µg dropped Super gently resuspending |
| 10µg Super gently resuspending Super | 20µg dropped Super gently resuspending 1:10 dilution | control |
2. Plate I(A is up)
| 10µg dropped Super | 10µg dropped Super gently resuspending | 10µg dropped Super gently resuspending |
| 10µg Super gently resuspending Super | 20µg dropped Super gently resuspending 1:10 dilution | control |
3. Plate I(A is up)
| 10µg dropped Pellet | 10µg dropped Pellet gently resuspending | 10µg dropped Pellet gently resuspending |
| 10µg Super gently resuspending Pellet | 20µg dropped Pellet gently resuspending 1:10 dilution | control |
The results
We only took some pictures from single cells, because there was no significant difference in fluorescence between each sample.
The main conclusion of this experiment is, that we have to get the flow cytometry started.
4.7 Transfection with different amounts of plasmids
The motivation
We want to check if the mVenus expression and transduction efficiency can be improved with higher amounts of DNA.
The plan
| Days | 4.7 | 5.7 | 6.7 | 7.7 | 8.7 | 9.7 | 10.7 | 11.7 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | FACS |
The Transfection was performed with 10, 18 and 24 µg per plasmid (mVenus, pHelper and R/C).
The virus harvest was performed according to standard protocol:
- 3 Plates got transduced
1. Plate I(A is up) completely 10 µg
| 300µl resuspended | 300µl resuspended | 450µl resuspended |
| 600µl resuspended | 600µl resuspended | control |
2. Plate II(A is up)completely 18 µg
| 300µl resuspended | 300µl resuspended | 450µl resuspended |
| 600µl resuspended | 600µl resuspended | control |
3. Plate III(A is up) completely 24 µg
| 300µl resuspended | 300µl resuspended | 450µl resuspended |
| 600µl resuspended | 600µl resuspended | control |
The results
FACS analysis of different treated HT1080 cells was done (18 samples)
The first percentage refers to the living and fluorescent cells and the second percentage to the total amout of living cells.
The data are listed this way:
1. Plate I(A is up)
| A1 | A2 | A3 |
| B1 | B2 | B3 |
1= Plate 1 A1 : 0% YFP-positive Cells (there were only a few cells in this sample)
2= Plate 1 A2 : 27,7% YFP-positve Cells from 73,3%
3= Plate 1 A3 : 17,3% " from 63,1%
4= Plate 2 A1 : 32,9% " from 75,3%
5= Plate 2 A2 : 27,2% " from 76,5%
6= Plate 2 A3 : 30,7% " from 73,4%
7= Plate 3 A1 : 26,8% " from 76,0%
8= Plate 3 A2 : 30,5% " from 74,2%
9= Plate 3 A3 : 30,7% " from 75,2%
10= Plate 1 B1 : 6,25% " from 36,1%
11= Plate 1 B2 : 6,42% " from 68,4%
12= Plate 1 B3 : 0,05% " from 89,5%
13= Plate 2 B1 : 20,5% " from 72,5%
14= Plate 2 B2 : 15,7% " from 73,6%
15= Plate 2 B3 : 0,02% " from 85,4%
16= Plate 3 B1 : 19,5% " from 73,3%
17= Plate 3 B2 : 20,8% " from 75,7%
18= Plate 3 B3 : 0,07% " from 84,8%
Conclusions
- Higher amounts of Plasmids (24, 30µg) seems to have no obvious effect on YFP expression
- Theres no difference in YFP expression comparing resuspended samples to not resuspended samples
- The amount of dead HT cells is about 25%, it is not clear if it is either AAV induced or mechanic stress (Trypsin, washing procedure)
Next steps:
- does the amount of GOI (ng plasmids) has an effect on YFP expression
- compare transduced to non transduced cells (vitality), or in other words is the mechanic stress responsible for 25% dead cell ratio?
- comparison of transduction efficiency of different virus stock production routines
14.7 Seeding AAV293 for TKGMK constructs
The motivation
We want to create the first viral particles with TKGMK as gene of interest.
The plan
| Days | 14.7 | 15.7 | 16.7 | 17.7 | 18.7 | 19.7 | 20.7 | 21.7 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | FACS |
30µg Plasmid were used (pHelper + pAAV_RC+ pAAV_RFC25_mgmk_TK30)
VIRUS no GOI :pHelper + pAAV_RC
VIRUS 1 :pHelper + pAAV_RC+ pAAV_RFC25_mgmk_TK30 clone 1 : P54
VIRUS 2 :pHelper + pAAV_RC+ pAAV_RFC25_mgmk_TK30 clone 2 : P55
VIRUS 3 :pHelper + pAAV_RC+ pAAV_RFC25_mgmk_TK30 clone 3 : P56
The results
Unfortunately the stocks never were used (exception: see following experiment, 7. august) because later on we noticed that the TKGMK plasmids were not in line with the RFC.
7.8 Seeding AAV293 for TKGMK and mVenus stocks
The motivation
After proofing the successfull transduction of tumor cells with the reportergen mVenus the next step is to kill the cells with a prodrug approach. The prodrug of our choice is the guanosine anolog ganciclovir, once activated ganciclovir gets integrated into the growing dna chain which results in termination of dna synthesis and cell death. We transduce the tumor cell line HT1080 with viral particles packed with the thymidine kinase guanosine mono phosphate kinase fusion protein (TKGMK) as gene of interest and the incubation with ganciclovir should kill the cells.
The plan
| Days | 7.8 | 8.8 | 9.8 | 10.8 | 11.8 | 12.8 | 13.8 | 14.8 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | FACS and microscopy |
| Plate | pAAV_RC/µg | pHelper/µg | GOI/µg |
| 1 | 3,3 | 3,3 | no GOI |
| 2 | 6,6 | 3,3 | no GOI |
| 3 | 3,3 | 3,3 | 3,3 TK_GMK clone1 (P54) |
| 4 | 3,3 | 3,3 | 3,3 TK_GMK clone1 (P54) |
| 5 | 3,3 | 3,3 | 3,3 TK_GMK clone2 (P55) |
| 6 | 3,3 | 3,3 | 3,3 YFP |
| 7 | 3,3 | 3,3 | 10 YFP |
| 8 | 3,3 | 3,3 | 20 YFP |
| 9 | 3,3 | 3,3 | 40 YFP |
| 10 | 3,3 | 3,3 | 60 YFP |
The transduction was performed according to standard protocol.
Plate 1:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 1 | 300µl virus 2 |
| B | control, no virus | 600µl virus 1 | 600µl virus 2 |
Plate 2:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 3 | 300µl virus 4 |
| B | control, no virus | 600µl virus 3 | 600µl virus 4 |
Plate 3:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 5 | 300µl virus 6 |
| B | control, no virus | 600µl virus 5 | 600µl virus 6 |
Plate 4:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 7 | 300µl virus 8 |
| B | control, no virus | 600µl virus 7 | 600µl virus 8 |
Plate 5:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 9 | 300µl virus 10 |
| B | control, no virus | 600µl virus 9 | 600µl virus 10 |
Plate 6:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 3 | 300µl virus 4 |
| B | control, no virus | 600µl virus 3 | 600µl virus 4 |
Plate 7:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 3 | 300µl virus 4 |
| B | control, no virus | 500µl virus 3 | 500µl virus 4 |
Plate 8:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 300µl virus 5 | 300µl virus 5 |
| B | control, no virus | 600µl virus 5 | 600µl virus 5 |
The test for functionality for TK GMK was done with the Prodrug Cymeven ganciclovir 500mg from Roche. The final concentration was 0,05 mM in the wells. After two days pictures were taken.
The results
As you can see, the TKGMK cell killing mechanism works! The samples with targeted mVenus constructs couldnt be checked because the FACS machine was not working. The fluorescence was checked with microscopy, the transduction efficiency is still low.
14.8 Seeding different amounts of AAV293 for transfection
The motivation
We want to know which confluence of AAV293 cells is optimal for AAV production.
The plan
| Days | 14.8 | 15.8 | 16.8 | 17.8 | 18.8 | 19.8 | 20.8 | 21.8 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | FACS |
We seed different amounts of cells in 10 cm2, and check the highest titer.
| Plate | amount of seeded AAV-293 cells |
| 1 | 100 000 |
| 2 | 200 000 |
| 3 | 400 000 |
| 4 | 500 000 |
| 5 | 800 000 |
| 6 | 1 000 000 |
| 7 | 1 200 000 |
| 8 | 1 500 000 |
| 9 | 1 750 000 |
| 10 | 1 750 000 |
The cells were transfected with 40µg standard CMV_YFP, 3,3µg Rep/Cap and 3,3µg pHelper.
Transduction was performed according to standard protocol.
Plate 1 YFP: 200.000 cells per well
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 165µl virus (1) | 165µl virus (2) |
| B | control, no virus | 165µl virus (1) | 165µl virus (2) |
Plate 2 YFP: 200.000 cells per well
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 165µl virus (3) | 165µl virus (4) |
| B | control, no virus | 165µl virus (3) | 165µl virus (4) |
Plate 3 YFP: 200.000 cells per well
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 165µl virus (5) | 165µl virus (6) |
| B | control, no virus | 165µl virus (5) | 165µl virus (6) |
Plate 4 YFP: 200.000 cells per well
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 165µl virus (7) | 165µl virus (8) |
| B | control, no virus | 165µl virus (7) | 165µl virus (8) |
Plate 5 YFP: 200.000 cells per well
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 165µl virus (9) | 165µl virus (10) |
| B | control, no virus | 165µl virus (9) | 165µl virus (10) |
The results
The FACS was still not available but we could easily detect a significant higher YFP expression (70-80%) at the following positions:
- Plate 1: A2 and B2, A3 and B3
- Plate 2: A2 and B2, A3 and B3
Therefore we conclude that we have to seed between 100000 and 500000 cells for optimal virus production precedure.
17.8 transduction with viral stocks 20 µg and 40 µg GOI-plasmid
The Motivation
We want to FACS our stocks to get valid data for further investigation of optimal virus assembly. Therefore created stocks with 20 and 40 µg CMV_mVenus.
The Plan
| Days | 17.8 | 18.8 | 19.8 | 20.8 | 21.8 | 22.8 | 23.8 | 24.8 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | FACS |
| A | control no cells | 300 µl virus (40 µg YFP) | 300 µl virus (20 µg YFP) |
|---|---|---|---|
| B | control no virus | 500µl P38 (pAAV2_mVenus) | 1000µl P38 (pAAV2_mVenus) |
| A | control no cells | 300 µl virus (40 µg YFP) | 300 µl virus (20 µg YFP) |
|---|---|---|---|
| B | control no virus | 500µl P38 (pAAV2_mVenus) | 1000µl P38 (pAAV2_mVenus) |
| A | control no cells | 300 µl virus (40 µg YFP) | 300 µl virus (20 µg YFP) |
|---|---|---|---|
| B | control no virus | 500µl P38 (pAAV2_mVenus) | 1000µl P38 (pAAV2_mVenus) |
| A | control no cells | 300 µl virus (40 µg YFP) | 300 µl virus (20 µg YFP) |
|---|---|---|---|
| B | control no virus | 500µl P38 (pAAV2_mVenus) | 1000µl P38 (pAAV2_mVenus) |
The Results
Although we were told that the FACS would be available in time it wasn't. So the cells could unfortunately not be checked for their YFP expression =(.
16.8 Seeding AAV293 for transfection with new R/C
The motivation
We want examine the functionality of different R/C constructs and the new eGFP gene of interest. This transfection was performed with new AAV-293 cells which were thawed.
The plan
| Days | 16.8 | 17.8 | 18.8 | 19.8 | 20.8 | 21.8 | 22.8 | 23.8 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | FACS |
- 2x106 cells were seeded
- used plasmids:
- P50 c= 429 ng/µl (RC)
- P64 c= 500 ng/µl (pHelper)
- P229 c= 162 ng/µl (GOI=YFP)
- P228 c= 540 ng/µl (GOI=eGFP)
- P158a c= 1800ng/µl (RC mutant)
- P158a c= 1770ng/µl (RC mutant)
The transfection was performed according to standard protocol.
- 1. plate: Lipo-Transfection => named Lipo
- 2. plate: Lipo-Transfection => named Lipo
- 3. plate: 3,3 µg (RC), 3,3 µg (pHelper), 3,3 µg (YFP) => was not used for transduction
- 4. plate: 10 µg (RC), 10 µg (pHelper), 3,3 µg (YFP) => named CaCl2
- 5. plate: 10 µg (RC mutant P158a), 10 µg (pHelper), 3,3 µg (YFP)=> CaCl2 10 µg RC mutant P158a
- 6. plate: 10 µg (RC mutant P158b), 10 µg (pHelper), 3,3 µg (YFP)=> CaCl2 10 µg RC mutant P158b
- 7. plate: 3,3 µg (RC mutant P158a), 3,3 µg (pHelper), 3,3 µg (YFP)=> CaCl2 3,3 µg RC mutant P158a
- 8. plate: 3,3 µg (RC mutant P158b), 3,3 µg (pHelper), 3,3 µg (YFP)=> CaCl2 3,3 µg RC mutant P158b
- 9. plate: 10 µg (RC), 10 µg (pHelper), 3,3 µg (eGFP)=> CaCl2 eGFP 3,3 µg
- 10. plate: 10 µg (RC), 10 µg (pHelper), 10 µg (eGFP)=> CaCl2 eGFP 10 µg
- 11. plate: 10 µg (RC), 10 µg (pHelper), 20 µg (eGFP)=> CaCl2 eGFP 20 µg
- 12. plate: 10 µg (RC), 10 µg (pHelper), 40 µg (eGFP)=> CaCl2 eGFP 40 µg
The results
As you can see the eGFP constructs worked very well. And even the transduction efficiency of the standard R/C is ok, the conclusion is that in previous transfections the AAV293 cells were to confluent so their transfection capability. The confluence of the cells is crucial! It also seems that our used amounts of plasmids had no decisive effect on the YFP expression because of the obviously variating results. A final conclusion is still due.
19.8 Seeding AAv293 for transfection with different HBS buffers
The Motivation
We want to try different 2xHBS Buffers (pH) and their influence on transfection efficiency!
The Plan
| Days | 19.8 | 20.8 | 21.8 | 22.8 | 23.8 | 24.8 | 25.8 | 26.8 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | FACS |
We try 5 different 2xHBS buffers:
- pH: 7.06
- pH: 7.08
- pH: 7.10
- pH: 7.12
- pH: 7.14
Used Plasmids: 10µg of each plasmid
- GOI: mVENUS => P262, conc:1148ng/µl; used amount: 8,71µl
- GOI: TKGMK => P82b, conc:1500ng/µl; used amount: 6,67µl
- pHELPER => P64, conc:503ng/µl; used amount: 20µl
- RepCap => 10.8: 1348ng/µl; used amount: 7,41µl
- Transduction of three 6-well plates:
Plate 1 YFP: 150.000 cells per well
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 500µl virus (1) | 500µl virus (2) |
| B | control, no virus | 500µl virus (1) | 500µl virus (2) |
Plate 2 YFP: 150.000 cells per well
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 500µl virus (3) | 500µl virus (4) |
| B | control, no virus | 500µl virus (3) | 500µl virus (4) |
Plate 3 YFP: 200.000 cells per well
| 1 | 2 | 3 | |
|---|---|---|---|
| A | control, no cells | 500µl virus (5) | 500µl virus (9) |
| B | control, no virus | 500µl virus (9) | 500µl virus (5) |
- (1)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,06
- (2)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,08
- (3)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,112
- (4)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,12
- (5)= 10µg RC, 10µg pHelper, 10µg GOI (YFP); pH(2xHBS)=7,14
the Results
AAV-Harvesting is at 24.8, same day Transduction and FACS will be done at 26.8
As you can see the 2xHBS with pH 7.112 performed best, in future experiments this buffer will be used.
22.8: Transfection of AAV293 with different amounts of cells, and two plasmid concentrations
The motivation
We want to define the final amount of AAV293 per 10 cm2 for optimal AAV-Production
The plan
| Days | 22.8 | 23.8 | 24.8 | 25.8 | 26.8 | 27.8 | 28.8 | 29.8 | 30.8 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest + Transduction | - | FACS |
The following amount of cells were seeded in 10 cm2:
- 100.000
- 100.000
- 200.000
- 200.000
- 300.000
- 300.000
- 400.000
- 400.000
- 500.000
- 500.000
- 600.000
- 600.000
- 700.000
- 700.000
10µg RC P158a four times mutant, 10µg pHelper, 3,3 µg mVenus P162 were pipetted on plates: 2, 4, 6, 8, 10 , 12, 14
3,3µg RC P158a four times mutant, 3,3µg pHelper, 3,3 µg mVenus P262 were pipetted on plates: 1, 3, 5, 7, 9, 11, 13
The results
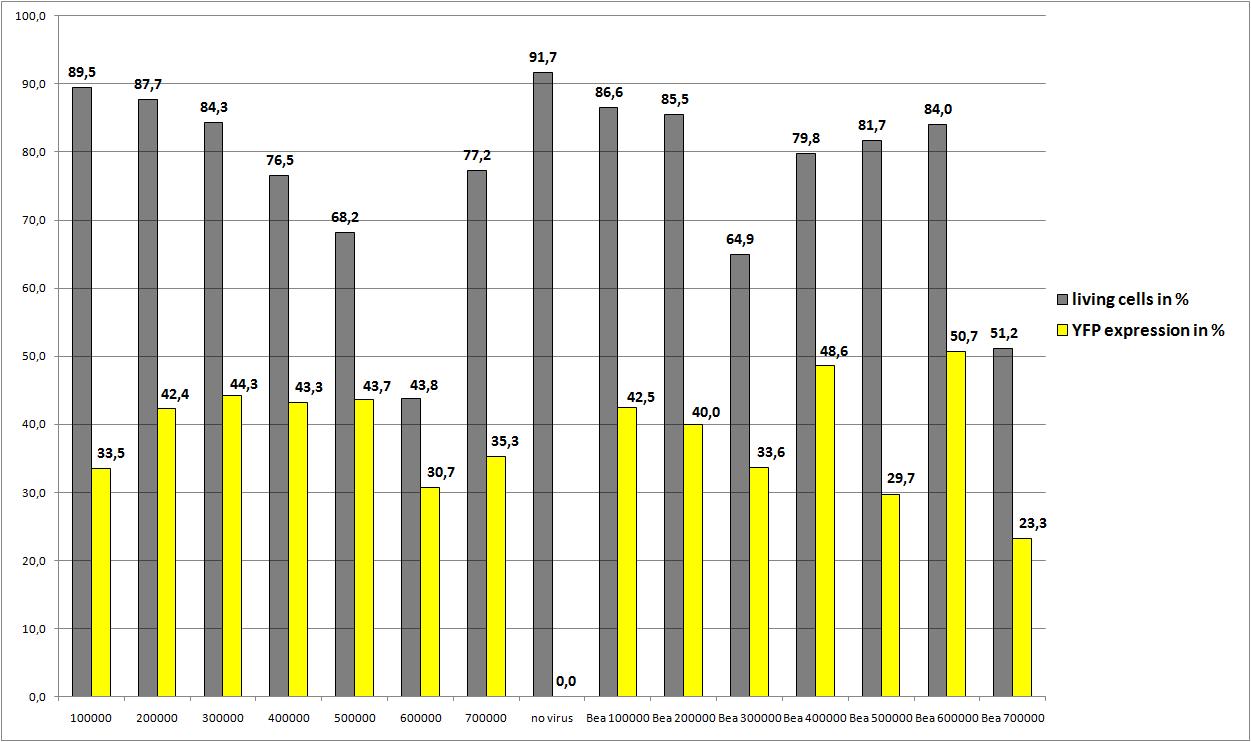
There is only one value for each approach but nevertheless the average YFP expression should be higher.
We really dont know the reason for the quite low YFP expression. There are also very good news: the reassembled construct (pSB1C3_leftITR_CMV_betaglobin_mVenus_hGH_rightITR) created from the single Biobricks works as "well" as the AAV2 with CMV_mVenus!
30.8 Seeding AAV293 for Transfection of 10 cm2 plates
The motivation
We want to create ten hypothetical identical viral stocks to estimate the standard deviation.
The plan
| Days | 30.8 | 31.8 | 1.9 | 2.9 | 3.9 | 4.9 | 5.9 | 6.9 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | FACS |
Transfection will be done with:
- RepCap:P357 10µg =>conc.:1274,8ng/µl used amount: 7,84µl
- pHelper:P356 10µg =>conc.:1068ng/µl used amount: 9,36µl
- mVenus:P261 => excel sheet P263: 10µg =>conc.:979ng/µl used amount: 10,21 µl
0,5 ml of each stock be stored at -80°C.
9,5 ml of each stock will be used for Transduction.
Result:
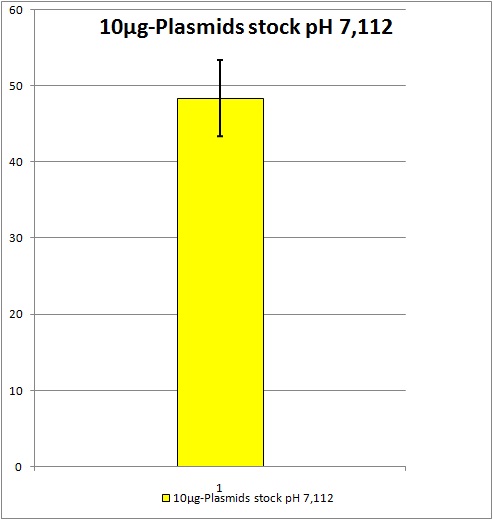
Standard deviation: about +/- 4,98% YFP expression. All future AAV2 stocks will be pooled if produced with this same plasmids and amounts of plasmids. Unfortunately the YFP expression is still located at around 50%. We would like to have a YFP expression of at least 70% because as soon as we start to test our not yet ready modified AAV2 we suggest the YFP expression will obviously decrease.
4.9 Seeding cells for checking constructs (GOIs) with/without HGH and beta globin and reassembled as control
The motivation
We want to check the functionality of these genes of interest
The plan
| Days | 4.9 | 6.9 | 7.9 | 8.9 | 9.9 | 10.9 | 11.9 | 12.9 |
|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | Seeding HT | Harvest + Transduction | FACS |
- P269 pSB1C3_lITR_CMV_beta-globin_mVenus_HGH_rITR = 325 ng/µl => 9,66 µl
- P270 pSB1C3_lITR_CMV_beta-globin_mVenus_HGH_rITR = 561 ng/µl => 7,26 µl
- P377 pSB1C3_lITR_CMV_mVenus_HGH_rITR = 325,04 ng/µl => 30,76 µl
- P378 pSB1C3_lITR_CMV_beta-globin_mVenus_rITR = 561 ng/µl => 17,8µl
Additionally P50: pAAV_RC R/C (26,45 µl) and pHELPER (14,61µl) pipetted to each approach.
The results
6.9 checking five ganciclovir concentrations for two day incubation approach optimal medication
The motivation
We want to check the optimal ganciclovir concentration, we use the TK/GMK Stocks (10 µg from each plasmid) from 24.8 with Buffer pH 7,10 and the pH12 stock.
The plan
| Days | 3.9 | 4.9 | 5.9 | 6.9 |
|---|---|---|---|---|
| Action | seeding HT | Transduction | - | FACS |
We want to use five different ganciclovir concentrations (the concentrations are absolute to the volume in the wells):
- 48.5µM
- 97 µM
- 0.485 mM
- 0.97 mM
- 4.85 mM
the results
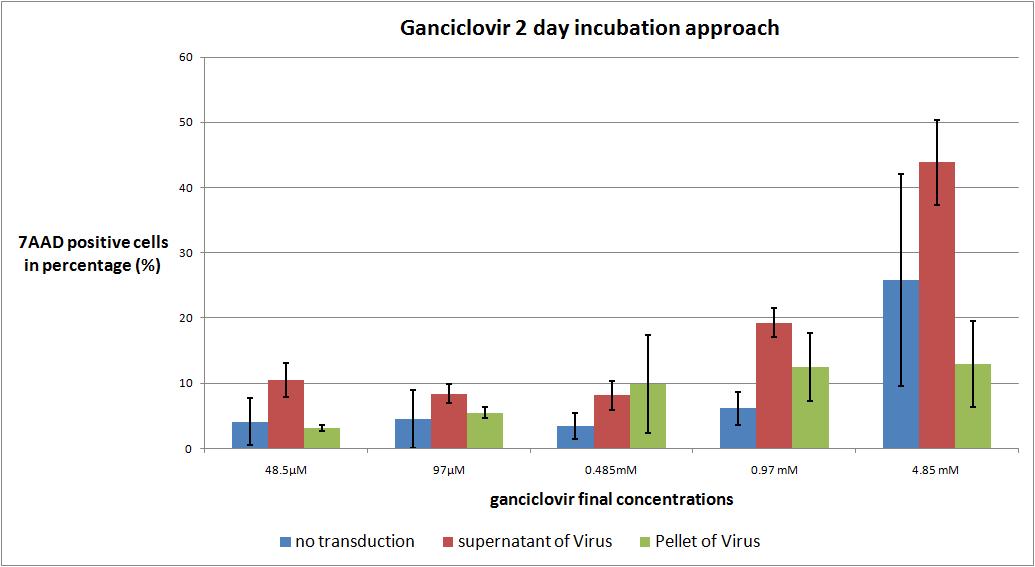
As expected the highest concentration of ganciclovir worked the best but it is remarkable that there is no significant difference between the first three ganciclovir amounts. The most remarkable note is, that in future experiments the incubation time has to be extended to 3 or 4 days.
15.9 Seeding AAV293 for testing our Affibody and other VP-mVenus constructs
The motivation
The pCerulean_VP1up_mVenus_Vp2/3 Construct will be tested in three different compositions.
The plan
We want to test these constructs:
- pCerulean_VP1up_NLS_Affibody_VP2/3
- pCerulean_VP1up_His_VP2/3
- pCerulean_VP1up_mVenus_Vp2/3
The His-tag is for purify our constructs and in this case we want to test, if the particles are still infectious. Construct with the affibody will be tested for functionality and even for their better affinity to our EGFR overexpressing cell line A431.
The results
Interpretation and discussion of our results.
19.9 Seeding AAV293 for checking two different Rep/Cap Constructs
The motivation
During the project 22 silent nucleotide exchange mutations were introduced into the capsid coding construct to make it compatible with the RFC standards and to have single cutting restriction enzymes flanking the 453 and the 587 loop sequence. Two point mutations had to be dismissed, because either a first test transduction showed that the construct was not working anymore or the insertion of the synthesized gene posed serious problems, because the restriction enzyme did not work.
The plan
Transfection and transduction was performed according to standard protocol.
The results
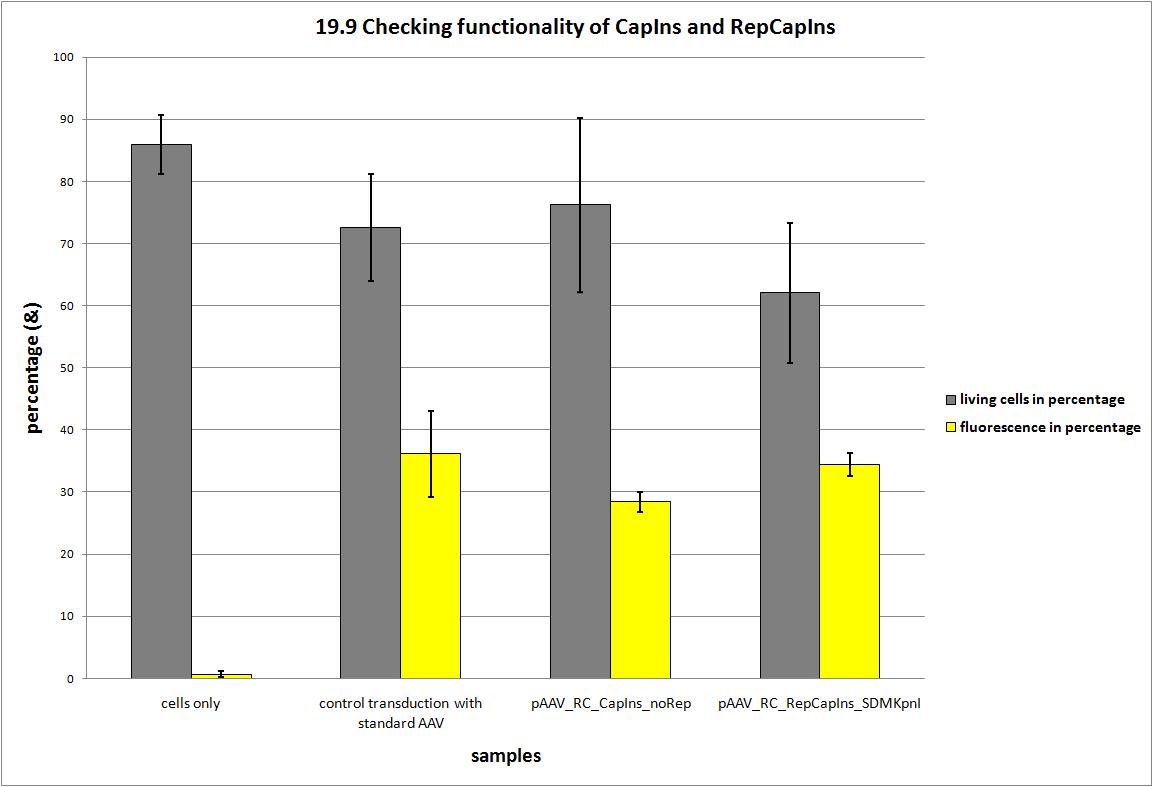
Finally, we have a construct that is shown to produce infectious particles comparable to the current AAV systems and carries 20 point mutations. Now we can announce that the Adeno-associated Virus is compatible to the RFC standard and the idea to replace the loop sequences via ViralBricks works!
22.9 Seeding AAV293 for transfection with different mVenus constructs for checking specific promoter activity
The motivation
We want to check if the CMV and pTERT promoters work equaly in the AAV293, A431 and HT1080 cells. So we transfect them via standard transfection CaCl2. It is obvious that the individual cell lines are not equaliy competent to transfection (the AAV293 are optimised for transfection). It seems that this promoter is not that active in AAV293 cells.
The plan
We transfected the three cell lines A431, AAV293 and HT1080 with following three constructs:
- CMV_mVenus
- CMV_eGFP
- pTERT_mVenus
Remember, that mVenus and eGFP have the exact same excitation wavelenght. We took the pictures with the same parameters, but saved the data in the wrong formate (.tif instead of .zvi), so we werent able to change the YFP expression from green to yellow.
| Days | 22.9 | 23.9 | 24.10 | 25.10 | 26.10 |
|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | fluorescence microscopy | fluorescence microscopy |
The results
As excepted, the expression of the CMV_mVenus and CMV_eGFP was very high even at 16h post transfection. The pTERT_mVenus construct worked aswell, but significantly weaker!
Problematic is the fact, that there was no expression in HT1080 and A431 detectable. Due the fact that these cell lines are not optimized for CaCl2 transfection. We can't evaluate if the bad transfection efficiency or the low promoter activity of pTERT are responsible for the results.
22.9 Testing of Hannas 6 VP-constructs with different compositions (10%, 25%, 50% and 75% fractions) with a VP2-knockout construct
The motivation
Test the following 18 constructs for functionality on A431 and HT1080 cells. Each of these constructs getting transfected in different compositions 25%, 50%, 75% and 90% with VP2_knockout 75%. In this way we are able to create viral particles with this ratios of VP proteins. We hope that the created particles are still infectious.
- pCerulean_CFP_MiddleLinker_VP2/3insCap P528
- pCerulean_Zegfr:1907_ShortlLinker_VP2/3insCap P527
- pCerulean_6xHis_Middlelinker_VP2/3insCap P534
- pCerulean_Zegfr:1907_Middlelinker_VP2/3insCap P535
- pCerulean_Zegfr:1907_SEG_VP2/3_Capins 10:90 P502
- pCerulean_Zegfr:1907_LongLinker_VP2/3_CapIns P503
The plan
| Days | 20.9 | 21.9 | 22.9 | 23.9 | 24.9 | 25.9 | 26.9 | 27.9 | 28.9 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest + Transduction | - | FACS |
The results
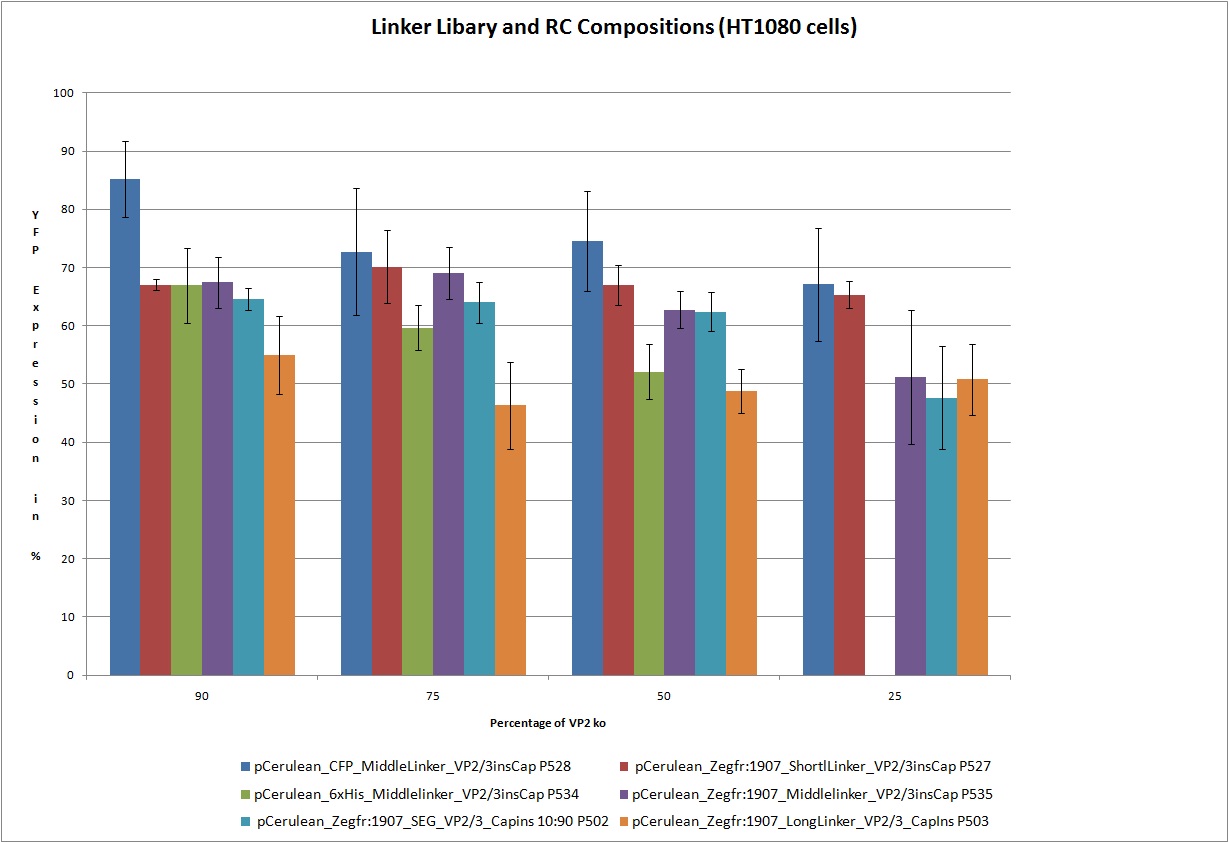
Obviously the constructs still work!
22.9 Seeding cells for testing pTERT_mVenus in R/C P326 and P431
The motivation
We want to check if the pTERT-promoter is realiable for tumor specific gene expression, we use two different kinds of R/C.
The plan
| Days | 22.9 | 23.9 | 24.9 | 25.9 | 26.9 | 27.9 | 28.9 | 29.9 | 30.9 | 1.10 | 2.10 | 3.10 | 4.10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest | - | - | Seeding HT, A293 and A431 | Transduction | FACS |
The results
29.9 Checking location of VP proteins via fluorescence microscopy and checking fractions of centrifugation steps via fluorescence spectrometer
The motivation: We want to figure out if it is possible to purify our viral stocks via centrifugation with 10.000 g and/or 20.000 g. In theory/ according to the literature the viral particles should be in the cells and attached to the HSPGs at the cellular surfaces.
The plan: Two different viral stocks were prepared:
Stock 1 with VP2-mVenus-fusion-Capsid loaded with TKGMK (4 wells from a 6 well plate)
- R/C 1: 50% pCerulean_VP1up_NLS_mVenus_Vp2/3 P501
- R/C 2: 50% Rep/Cap2: P449 pAAV_RC_4fachmut_VP1-ko:
- GOI: TKGMK: P82.b
- pHelper
Stock 2 "standard" virus (4 wells from a 6 well plate)
- R/C 1 P486
- GOI: TKGMK: P82.b
- pHelper
These two stocks got harvested according to standard protocol (scratching cells, transfer them in to 15 ml falcons, resuling 12ml solution total). After centrifugation (10 min 200g), the supernatant got transferred into two new 15 ml falcons and the pellets were resuspended with 10 ml DMEM Medium. The Stocks got freezed thawed two times so finally there were:
- DMEM as negative control
- resuspended Pellet with mVenus-fusioned viral capsids
- resuspended Pellet with "standard virus"
- supernatant with mVenus-fusioned viral capsids
- supernatant with "standard virus"
The fluorescence of each stock was measured via spectrometer. After that, stocks 2 and 4 were centrifugated 10min with 10.000 g followed by an other fluorescence measuring. Finally the stocks got spin down with 20.000 g for 10 min
The results:
The conclusions:
The highest amount of fluorescence was measured in the supernatant, the individual centrifugation steps had no decrease in fluorescence as consequence!
Keep in mind, without further investigation it is not valid to say that we can purify our stocks via 10-20.000 g centrifugation steps, because we dont know if the viral capsids are still intact! The FACS analysis are not sufficient enough to make a decision, because our last samples had YFP as GOI (=> so efficient cell sorting was not possible). We need to do qPCRs to make a valid evidence.
30.9 Production of new Standard mVenus Vector
The motivation
The AAV2 with 4 point mutations, inserted rep and cap and remutated KpnI restriction has to be checked for its functionality in order to compare it with the wild type AAV2. Hopefully there will be no significant difference.
The plan
After the "delay" 200.000 A431 and HT1080 cells were seeded and transduced on the following day with 0,5 ml of this AAV2 mutant.
| Days | 30.9 | 1.10 | 2.10 | 3.10 | 4.10 | 5.10 | 6.10 | 7.10 | 8.10 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest | delayed because of cell death | delayed because of cell death |
The results
The stocks were harvested successfully
30.9 Seeding AAV293 for production of standard TKGMK vector
The motivation
We want to create a big amount of viral particles with TKGMK as gene of interest for using it as an standard vector for the MTT-assay.
The plan
| Days | 30.9 | 1.10 | 2.10 | 3.10 | 4.10 | 5.10 | 6.10 | 7.10 | 8.10 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest | delayed because of cell death | delayed because of cell death |
The results
The viral stocks were harvested efficiently and are ready to use.
30.9 Seeding AAV293 for production of vectors with mVenus missing beta globin
The motivation
Beta globin is said to improve (stratagene) the expression so we expect a worse mVenus (reportergene) expression compared to the standard AAVs. Therefore the seeded A431 and HT1080 cells were transduced with a AAV2 mutant missing beta-globin.
The plan
Ater the "delay" 200.000 HT1080 and A431 cells were seeded and transduced with 0,5 ml AAV2 mutant on the following day.
| Days | 30.9 | 1.10 | 2.10 | 3.10 | 4.10 | 5.10 | 6.10 | 7.10 | 8.10 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest | delayed because of cell death | delayed because of cell death |
The results
Interpretation and discussion of our results.
30.9 Seeding AAV293 for production of vectors with mVenus missing HGH
The motivation
HGH is responsible for the mRNA polyadenylation of the virus so we expect expect a worse mVenus (reportergene) expression compared to the standard AAVs. Therefore the seeded A431 and HT1080 cells were transduced with a AAV2 mutant missing HGH.
The plan
After the "delay" 200.000 A431 and HT1080 cells were seeded and transduced on the following day with 0,5 ml AAV2 mutant missing HGH.
| Days | 30.9 | 1.10 | 2.10 | 3.10 | 4.10 | 5.10 | 6.10 | 7.10 | 8.10 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest | delayed because of cell death | delayed because of cell death |
The results
Interpretation and discussion of our results.
30.9 Seeding AAV293 for testing RC with HSPG knockout for functionality
The motivation
The primary AAV2 receptor (the HSPG receptor) was knocked out so we expect a worse tranduction rate (about 0,3 to 0,4 of the standard transduction rate, assuming that there are HSPGs on the HT1080 and A431 cell surface). The worse the better.
The plan
After the "delay" 200.000 A431 and HT1080 cells were seeded and transduced on the following day with 0,5 ml AAV2 mutant with a HSPG-receptor knockout.
| Days | 30.9 | 1.10 | 2.10 | 3.10 | 4.10 | 5.10 | 6.10 | 7.10 | 8.10 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest | delayed because of cell death | delayed because of cell death |
The results
There is a significant difference in transduction efficiency. The actual reduction is even higher than expected, according to literature it should be around 60%, we reached 80%. We measured via qPCR the amount of viral particles for each stock, and they are nearly the same the HSPG Knockout has 2*107particles per ml , the stock with HSPG-binding motif has 4.5*106 particles per ml.
30.9 Seeding AAV293 for testing RC WITHOUT HSPG knockout for functionality
The motivation
The produced vector has the intact HSPG-binding motif, so we can compare it with the vector produced in: "Seeding AAV293 for testing RC with HSPG knockout for functionality"
The plan
Transfection and transduction were performed according to standard protocol.
| Days | 30.9 | 1.10 | 2.10 | 3.10 | 4.10 | 5.10 | 6.10 | 7.10 | 8.10 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest | delayed because of cell death | delayed because of cell death |
The results
See 30.9 Seeding AAV293 for testing RC with HSPG knockout for functionality
12.10 Seeding AAV293 cells for transfection with VP-BAP
The motivation
We want to create a viral stock for purification and biotinylation, the ratio of the Rep/Cap plasmids is 1:1.
The plan
Transfection was performed according to standard protocol:
- Gene of interest: mVenus
- R/C: pSB1C3-001_pCMV_[AAV]VP123_587_BAP in cotransfection with pSB1C3_001_RC_IRCK_P5tataless clone 1
| Days | 12.10 | 13.10 | 14.10 | 15.10 | 16.10 | 17.10 | 18.10 | 19.10 | 20.10 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest + Transduction | - | FACS |
The results
12.10 Seeding AAV293 cells for transfection with mVenus as gene of interest and pCerulean_VP1up_NLS_mVenus_VP2/3_HSPG-KO
The motivation
We want to create a viral stock for purification and biotinylation, the ratio of the Rep/Cap plasmids is 1:1.
The plan
Transfection was performed according to standard protocol:
- Gene of interest: mVenus
- R/C: pCerulean_VP1up_NLS_mVenus_VP2/3_HSPG-KO in cotransfection with pSB1C3_001_RC_IRCK_VP1-ko_HSPG-ko_P5tataless cl1
| Days | 12.10 | 13.10 | 14.10 | 15.10 | 16.10 | 17.10 | 18.10 | 19.10 | 20.10 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest + Transduction | - | FACS |
The results
12.10 Seeding AAV293 cells for transfection with mVenus as GOI and CFP_Middlelinker_VP2/3_HSPG-KO
The motivation
This viral stock is for live imaging and probably for western blotting, the ratio of the Rep/Cap plasmids is 1:1.
The plan
| Days | 18.10 | 19.10 | 20.10 | 21.10 | 22.10 | 23.10 | 24.10 | 25.10 | 26.10 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest + Transduction | - | FACS |
Transfection was performed according to standard protocol:
- Gene of interest: mVenus
- R/C: CFP_Middlelinker_VP2/3_HSPG-KO in cotransfection with pSB1C3_001_RC_IRCK_P5tataless clone 1
The results
17.10 Production of AAV2 with 587-His
Investigator Patrick
The motivation
We wanted to produce AAV2 which could be purified via affinity purification. Therefore 10 x 10cm^2 were seeded with AAV-293. Five of these plates contained serum-free medium and cells accustomized to this.
Used plasmids:
- pHelper
- pSB1C3_leftITR_CMV_betaglobin_mvenus_hGH_rightITR (P541)
- 50% pSB1C3_001_RC_IRCK_P5tataless (P712)
- 50% pSB1C3-001_pCMV_[AAV2]VP123_587_HIS (P638)
The plan
In this section, the design of the experiment is explained.
| Days | 17.10 | 19.10 | 20.10 | 21.10 | 22.10 | 23.10 | 24.10 | 25.10 | 26.10 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | Transfection | - | Seeding HT | AAV2 harvest | Transduction | FACS |
The results
Interpretation and discussion of our results.
18.10 Seeding AAV293 cells for transfection Superconstructs
The motivation
We want to create a viral stock for purification and biotinylation, the ratio of the Rep/Cap plasmids is 1:1.
The plan
| Days | 18.10 | 19.10 | 20.10 | 21.10 | 22.10 | 23.10 | 24.10 | 25.10 | 26.10 |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest + Transduction | - | FACS |
Transfection was performed according to standard protocol:
Gene of interest: CMV_mVenus
- pSB1C3_CMV_Zegfr:1907_MiddleLinker_VP2/3_587-KO_6xHis in ratio 1:1 pSB1C3_001_RC_IRCK_VP2-ko_HSPG-ko_P5tataless cl1
- pSB1C3-001_pCMV_[AAV2]VP123_453_Z34C in ratio 1:1 pSB1C3_001_RC_IRCK_HSPG-ko_P5tataless_RFC10
- pSB1C3_001_CMV_VP123_587-KO_Z34C in ratio 1:1 pSB1C3_001_RC_IRCK_HSPG-ko_P5tataless_RFC10
- pSB1C3_001_CMV_VP123_587-KO_Z34C_spacer in ratio 1:1 pSB1C3_001_RC_IRCK_HSPG-ko_P5tataless_RFC10
Gene of interest: pTERT_TKGMK
- pSB1C3_CMV_Zegfr:1907_MiddleLinker_VP2/3_587-KO_6xHis in ratio 1:1 pSB1C3_001_RC_IRCK_VP2-ko_HSPG-ko_P5tataless cl1
Gene of interest: CMV_TKGMK
- pSB1C3_CMV_Zegfr:1907_MiddleLinker_VP2/3_587-KO_6xHis in ratio 1:1 pSB1C3_001_RC_IRCK_VP2-ko_HSPG-ko_P5tataless cl1
The results
18.10 Seeding AAV293 cells for transfection with Cytosindeaminase
The motivation
We want to create viral stocks with CMV_Cytosindeaminase and pTERT_Cytosindeaminase as gene of interests, to check qualitatively the bystander effect.
The plan
| Days | 18.10 | 19.10 | 20.10 | 21.10 | 22.10 | 23.10 | 24.10 | 25.10 | 26.10 | 27.10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | - | Transfection | - | - | Seeding HT | Harvest + Transduction | - | FACS |
Transfection, harvest and transduction were performed according to standard protocol:
Gene of interest: CMV_Cytosindeaminase
- P712
Gene of interest: pTERT_Cytosindeaminase
- P712
The results
22.10.2010 Transduction with old mVenus Stocks for taking pictures
The motivation
We transduced HT1080 with our stocks packaged with CMV_mVenus.
The plan
| Days | 22.9 | 23.9 | 24.9 | 25.9 |
|---|---|---|---|---|
| Action | seeding HT1080 | Transduction | - | Fluorescence microscopy |
The results
23.10 Testing the bystander effect of the cytosine deaminase
The motivation
The activated form of 5-fluorocytosine has the ability to diffuse through the plasma membrane. In this way cells get killed which were not transduced by the viral particles. We use an old yfp stock t determine if the transgene expression is on and to harvest the cells which are transduced with cytosine deaminase loaded AAV2.
The plan
| Days | 23.10 | 24.10 | 25.10 | 26.10 | 27.10 | 28.10 |
|---|---|---|---|---|---|---|
| Action | Transduce HT1080 and seed HT1080 | Transfer the transduced HT1080 onto the non transduced HT1080 | - | - | - | Measure living cells via Neubauer cellchamber |
Gene of interest: CMV_Cytosindeaminase
- P712
Gene of interest: CMV_mVenus
- P468
All dishes had this order:
| 1 | 2 | 3 | |
|---|---|---|---|
| A | Medium with prodrug | Medium with prodrug | Medium with prodrug |
| B | Medium | Medium | Medium |
The concentration of the prodrug 5-fluorocytosine is 54 mmolar
Cells were seeded following this scheme:
plate 1
| 1 | 2 | 3 | |
|---|---|---|---|
| A | 1*105 HT1080 transduced with CMV_Cytosindeaminase | 1*105 HT1080 transduced with CMV_Cytosindeaminase | 1*105 HT1080 transduced with CMV_Cytosindeaminase |
| B | 1*105 HT1080 transduced with CMV_Cytosindeaminase | 1*105 HT1080 transduced with CMV_Cytosindeaminase | 1*105 HT1080 transduced with CMV_Cytosindeaminase |
plate 2
| 1 | 2 | 3 | |
|---|---|---|---|
| A | 2*105 HT1080 non transduced | 2*105 HT1080 non transduced | 2*105 HT1080 non transduced |
| B | 2*105 HT1080 non transduced | 2*105 HT1080 non transduced | 2*105 HT1080 non transduced |
The next plates were seeded with non transduced cells and 1*105 of the cytosin deaminase transduced cells
plate 3
| 1 | 2 | 3 | |
|---|---|---|---|
| A | 2*105 HT1080 | 2*105 HT1080 | 2*105 HT1080 |
| B | 2*105 HT1080 | 2*105 HT1080 | 2*105 HT1080 |
The results
25.10.2010 Transduction with several constructs for Flow Cytometry
The motivation
We want to quantify via flow cytometry the Affibody superconstruct, Darpin HSPG-KO Construct and standard R/C with HSPG-KO.
The plan
| Days | 25 | 26 | 27 |
|---|---|---|---|
| Action | Transduction | - | FACS |
The results
BLANK EXPERIMENT
The motivation
The plan
| Days | - | - | - | - | - | - | - | - | - |
|---|---|---|---|---|---|---|---|---|---|
| Action | seeding AAV293 | - | Transfection | - | - | Seeding HT | Harvest + Transduction | - | FACS |
The results
Citations
[1] Media:Freiburg10_Stratagene_AAV_helper_free_manual.pdf
[2] [http://www.copewithcytokines.de/cope.cgi?key=A431 Titel einfügen]
[3] Read, A. P.; Strachan, T. (1999). "Chapter 18: Cancer Genetics". Human molecular genetics 2. New York: Wiley. ISBN 0-471-33061-2.
[4] [http://www.biomol.de/dateien/infos_nr778.gif Titel einfügen]
[5] Media:Freiburg10 Titration of AAV-2 particles via a novel capsid ELISA.pdf
 "
"