Team:Freiburg Bioware/NoteBook/Labjournal/June
From 2010.igem.org
| (13 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
{{:Team:Freiburg_Bioware/Head}} | {{:Team:Freiburg_Bioware/Head}} | ||
{{:Team:Freiburg_Bioware/menu_notebook}} | {{:Team:Freiburg_Bioware/menu_notebook}} | ||
| + | {{:Team:Freiburg_Bioware/jquery}} | ||
<!-- Freiburg_bioware --> | <!-- Freiburg_bioware --> | ||
| Line 9: | Line 10: | ||
<left><u1>NoteBook Navigator</u1></left> | <left><u1>NoteBook Navigator</u1></left> | ||
<br> | <br> | ||
| - | |||
<ul> | <ul> | ||
<li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/March">March (labday 1)</a></li> | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/March">March (labday 1)</a></li> | ||
| Line 20: | Line 20: | ||
<li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/September">September part 1 (labday 107 - 123)</a></li> | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/September">September part 1 (labday 107 - 123)</a></li> | ||
<li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/September2">September part 2 (labday 124 - 135)</a></li> | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/September2">September part 2 (labday 124 - 135)</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/October">October part 1 (labday 136 - | + | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/October">October part 1 (labday 136 - 149 )</a></li> |
| - | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/October2">October part 2 (labday | + | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/October2">October part 2 (labday 150 - 166 )</a></li> |
| - | + | ||
<li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/November">November (labday 167 - 170 )</a></li> | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/November">November (labday 167 - 170 )</a></li> | ||
| - | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/Cellculture">Cellculture </a></li> | + | <li><a href="https://2010.igem.org/Team:Freiburg_Bioware/NoteBook/Labjournal/Cellculture">Cellculture</a></li> |
</ul> | </ul> | ||
</div> | </div> | ||
| Line 123: | Line 122: | ||
==== <p style="font-size:17px; background-color:#00dd77;">19. Labortag 02.06.2010:</p>==== | ==== <p style="font-size:17px; background-color:#00dd77;">19. Labortag 02.06.2010:</p>==== | ||
| - | =====<p style="font-size:17px; background-color:#66bbff;"> | + | =====<p style="font-size:17px; background-color:#66bbff;">Picking clones of pAAV_RFC25MCS</p>===== |
<p><b>Investigators: Adrian, Bea, Chris W., Hanna, Anissa</b></p> | <p><b>Investigators: Adrian, Bea, Chris W., Hanna, Anissa</b></p> | ||
'''Practical work:''' <br> | '''Practical work:''' <br> | ||
| Line 129: | Line 128: | ||
4 colonies were picked and grown @ 37°C over night. | 4 colonies were picked and grown @ 37°C over night. | ||
<br> | <br> | ||
| - | + | ||
| + | =====<p style="font-size:17px; background-color:#66bbff;">Design and ordering of Oligos (NotI)</p>===== | ||
| + | <p><b>Investigators: Adrian, Bea, Chris W., Hanna, Anissa</b></p> | ||
Oligos for site directed mutagenesis of the NotI restriction sites in pAAV_MCS (ITRs) were designed: <br /> | Oligos for site directed mutagenesis of the NotI restriction sites in pAAV_MCS (ITRs) were designed: <br /> | ||
| - | [[ | + | [[Media:Freiburg10 NotI ITR Oligos.pdf]] |
<br> | <br> | ||
| Line 303: | Line 304: | ||
Plasmids received: <br> | Plasmids received: <br> | ||
| - | *'''pXX6''' | + | *'''pXX6''' all Adenovirus helper genes without AAV genes |
| - | *'''pKEX-2XL.Rep40''' | + | *'''pKEX-2XL.Rep40''' expression plasmid for Rep protein 40 |
| - | *'''pKEX-2XL.Rep 52''' | + | *'''pKEX-2XL.Rep 52''' expression plasmid for Rep protein 52 |
| - | *'''pKEX-2XL.Rep 68''' | + | *'''pKEX-2XL.Rep 68''' expression plasmid for Rep protein 68 |
| - | *'''pKEX-2XL.Rep 78''' | + | *'''pKEX-2XL.Rep 78''' expression plasmid for Rep protein 78 |
| - | *'''pCMV-VP(HS)''' | + | *'''pCMV-VP(HS)''' expression plasmid for VP proteins in combination (assemly competent) |
| - | *'''pKEX-VP1''' | + | *'''pKEX-VP1''' expression plasmid for single VP proteins (Cap: VP1, VP2 und VP3). Be careful: Single constructs are not assembly competent; Ruffing et al 1992; Steinbach et al. 1997; |
| - | *'''pKEX-VP2''' | + | *'''pKEX-VP2'''expression plasmid for single VP proteins (Cap: VP1, VP2 und VP3). Be careful: Single constructs are not assembly competent; Ruffing et al 1992; Steinbach et al. 1997; |
| - | *'''pKEX-VP3''' | + | *'''pKEX-VP3'''expression plasmid for single VP proteins (Cap: VP1, VP2 und VP3). Be careful: Single constructs are not assembly competent; Ruffing et al 1992; Steinbach et al. 1997; |
| - | *'''pTRUF_CMV_eGFP''' | + | *'''pTRUF_CMV_eGFP'''single stranded vector for eGFP expression |
| - | *'''dsAAV_CMV_eGFP'''"self complementary" | + | *'''dsAAV_CMV_eGFP'''"self complementary" expression vector from X. Xiao |
| - | *'''pTAV2 ( | + | *'''pTAV2 (whole AAV genome in "blue script" vector) |
| - | *'''pDG''' | + | *'''pDG''' complete helper plasmid, nedded for vector production ; Grimm et al. 1998 |
<br> | <br> | ||
*In order to obtain the DNA following steps have been performed: | *In order to obtain the DNA following steps have been performed: | ||
| Line 626: | Line 627: | ||
<u>Glycerol Stocks</u> | <u>Glycerol Stocks</u> | ||
{| border="1" | {| border="1" | ||
| - | | align="left" | ||align="left"| '''Clone 1''' ||align="left"| '''Clone 2''' ||align="left"| '''Clone 3''' ||align="left"| '''Clone 4''' ||align="left"| '''Clone 5''' ||align="left"| '''Clone 6''' ||align="left"| '''Clone 7''' ||align="left"| '''Clone 8''' ||align="left"| '''Clone 9 | + | | align="left" | ||align="left"| '''Clone 1''' ||align="left"| '''Clone 2''' ||align="left"| '''Clone 3''' ||align="left"| '''Clone 4''' ||align="left"| '''Clone 5''' ||align="left"| '''Clone 6''' ||align="left"| '''Clone 7''' ||align="left"| '''Clone 8''' ||align="left"| '''Clone 9''' |
|- | |- | ||
| - | | align="left" | '''Bacteria strain''' | + | | align="left" | '''Bacteria strain''' ||align="left"| XL1B ||align="left"| XL1B ||align="left"| XL1B ||align="left"| XL1B ||align="left"| XL1B ||align="left"| XL1B ||align="left"| XL1B ||align="left"| XL1B ||align="left"| XL1B |
|- | |- | ||
| - | | align="left" | '''Plasmidname''' ||align="left"| pXX6 ||align="left"| pKEX-2XL.Rep 40 ||align="left"| pKEX-2XL.Rep 52 ||align="left"| pKEX-2XL.Rep 68 ||align="left"| pKEX-2XL.Rep 78 ||align="left"| pCMV-VP(HS) ||align="left"| pKEX-VP1 ||align="left"| pKEX-VP2 ||align="left"| pKEX-VP3 | + | | align="left" | '''Plasmidname''' ||align="left"| pXX6 ||align="left"| pKEX-2XL.Rep 40 ||align="left"| pKEX-2XL.Rep 52 ||align="left"| pKEX-2XL.Rep 68 ||align="left"| pKEX-2XL.Rep 78 ||align="left"| pCMV-VP(HS) ||align="left"| pKEX-VP1 ||align="left"| pKEX-VP2 ||align="left"| pKEX-VP3 |
|- | |- | ||
| - | | align="left" | '''Date''' ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 | + | | align="left" | '''Date''' ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 |
|- | |- | ||
| - | | align="left" | '''given number''' ||align="left"| B14 ||align="left"| B15 ||align="left"| B16 ||align="left"| B17 ||align="left"| B18 ||align="left"| B19 ||align="left"| B20 ||align="left"| B21 ||align="left"| B22 ||align="left"| B23 ||align="left"| B24 ||align="left"| B25 ||align="left"| B26 | + | | align="left" | '''given number''' ||align="left"| B14 ||align="left"| B15 ||align="left"| B16 ||align="left"| B17 ||align="left"| B18 ||align="left"| B19 ||align="left"| B20 ||align="left"| B21 ||align="left"| B22 |
| + | |} | ||
| + | {| border="1" | ||
| + | | align="left" | ||align="left"| '''Clone 10''' ||align="left"| '''Clone 11''' ||align="left"| '''Clone 12''' ||align="left"| '''Clone 13''' | ||
| + | |- | ||
| + | | align="left" | '''Bacteria strain''' ||align="left"| XL1B ||align="left"| XL1B ||align="left"| XL1B ||align="left"| XL1B | ||
| + | |- | ||
| + | | align="left" | '''Plasmidname''' ||align="left"| pTRUF_CMV_eGFP ||align="left"| dsAAV_CMV_eGFP ||align="left"| pTAV2||align="left"| pDG | ||
| + | |- | ||
| + | | align="left" | '''Date''' ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 ||align="left"| 07.06.2010 | ||
| + | |- | ||
| + | | align="left" | '''given number''' ||align="left"| B23 ||align="left"| B24 ||align="left"| B25 ||align="left"| B26 | ||
|} | |} | ||
| Line 654: | Line 666: | ||
* Always check volumes - try to estimate if volume makes sense (check pipettes!) | * Always check volumes - try to estimate if volume makes sense (check pipettes!) | ||
* Don't discard bacteria cultures until glycerol stocks and mini-preps are successfully done!!! | * Don't discard bacteria cultures until glycerol stocks and mini-preps are successfully done!!! | ||
| - | <p style="font-size: | + | <p style="font-size:15px; font-weight: bold; color: red;">'''Today's conclusion: Better ask 2 times than do something wrong without asking!!!'''</p> |
<br> | <br> | ||
| Line 791: | Line 803: | ||
<b>Investigators: Jessy, Achim, Sven, Toby, Hanna</b> | <b>Investigators: Jessy, Achim, Sven, Toby, Hanna</b> | ||
<br> | <br> | ||
| - | <p style="font-size:15px; font-weight: bold; color: red;">'''No pAAV_iGEM-MCS_w/oPstI transformed bacteria (site-directed mutagensis) grew on the ampicillin agar plates over night! '''</p> Experiment is conducted again by Toby (Thanks a lot!!!).<br> | + | <p style="font-size:15px; font-weight: bold; color: red;">'''No pAAV_iGEM-MCS_w/oPstI transformed bacteria (site-directed mutagensis) grew on the ampicillin agar plates over night! '''</p> Experiment is conducted again, guided by Toby (Thanks a lot!!!).<br> |
<br> | <br> | ||
| - | <p style="font-size:15px; font-weight: bold; color: red;">'''Further on the cloning of mVenus-YFP into pAAV_iGEM-MCS seemed to fail: '''</p> Only on the agar plate cotaining the bacteria transformed with the "vector_oben" ligation, which actually should be the "wrong" ligation (gelextraction of a band which contained fragments that were too large, see picture in lab journal), revealed colonies. Therefore this experiments is also conducted one more time by Sven (thanks also a lot!!!)<br> | + | <p style="font-size:15px; font-weight: bold; color: red;">'''Further on the cloning of mVenus-YFP into pAAV_iGEM-MCS seemed to fail: '''</p> Only on the agar plate cotaining the bacteria transformed with the "vector_oben" ligation, which actually should be the "wrong" ligation (gelextraction of a band which contained fragments that were too large, see picture in lab journal), revealed colonies. Therefore this experiments is also conducted one more time, guided by Sven (thanks also a lot!!!)<br> |
<br> | <br> | ||
<p style="font-size:15px; font-weight: bold; color: fuchsia;"><u>Insertion of mVenus_YFP into pAAV_iGEM-MCS</u></p> | <p style="font-size:15px; font-weight: bold; color: fuchsia;"><u>Insertion of mVenus_YFP into pAAV_iGEM-MCS</u></p> | ||
*experiment date: 9.6.2010 | *experiment date: 9.6.2010 | ||
| - | + | ||
*plasmid: | *plasmid: | ||
**Vector: name: pAAV_iGEM-MCS number: 34.2 production date: 5.6.2010 | **Vector: name: pAAV_iGEM-MCS number: 34.2 production date: 5.6.2010 | ||
| Line 1,357: | Line 1,369: | ||
<b>Investigators: Achim, Hanna</b> | <b>Investigators: Achim, Hanna</b> | ||
| - | In order to sequence the hGH (polyadenylation) sequence and in general the GOI (gene of interest :) ) primers were designed: [[ | + | In order to sequence the hGH (polyadenylation) sequence and in general the GOI (gene of interest :) ) primers were designed: [[Media:Primer GOI+polyA pAAV iGEM-MCS mVenus YFP.pdf]] |
<br> | <br> | ||
These primer will be used in order to sequence P38 (pAAV_iGEM_mVenus-YFP) which delivered fluorescencing HT1080 cells - in comparison to the already sequenced P39 clone. Further on they can be used in general for sequencing any GOI. <br> | These primer will be used in order to sequence P38 (pAAV_iGEM_mVenus-YFP) which delivered fluorescencing HT1080 cells - in comparison to the already sequenced P39 clone. Further on they can be used in general for sequencing any GOI. <br> | ||
| Line 1,381: | Line 1,393: | ||
<br> | <br> | ||
| - | ====<p style="font-size:15px; background-color:#66bbff;">Picking clones of pAAV_RFC25 | + | ====<p style="font-size:15px; background-color:#66bbff;">Picking clones of pAAV_RFC25</p>==== |
<b>Investigators: Achim, Chris W., Hanna</b><br> | <b>Investigators: Achim, Chris W., Hanna</b><br> | ||
| - | Bacteria cultures are stored | + | Clones were picked, 250 mL DYT were inoculated.<br/> |
| + | Bacteria cultures are stored at 37°C over night. | ||
<br> | <br> | ||
| + | ====<p style="font-size:15px; background-color:#66bbff;"> Transduction results </p>==== | ||
<b>Investigators: Adrian (guided by Sven)</b> | <b>Investigators: Adrian (guided by Sven)</b> | ||
<gallery widths=400px heights=200px perrow=2 caption="Results of transduction, ~ 1% of the HT1080 showed expression of mVenus."> | <gallery widths=400px heights=200px perrow=2 caption="Results of transduction, ~ 1% of the HT1080 showed expression of mVenus."> | ||
| Line 1,411: | Line 1,425: | ||
Benzonase (will be used fur viral stock preparation) | Benzonase (will be used fur viral stock preparation) | ||
<br> | <br> | ||
| - | Primer for for ITR- | + | Primer for for ITR-conversion to iGEM-standars |
| - | *Presentations of Adrian and Patrick: [[ | + | *Presentations of Adrian and Patrick: [[Media:Adrian.ppt]] [[Media:Patrick.ppt]] |
<br> | <br> | ||
Latest revision as of 21:36, 27 October 2010
- March (labday 1)
- April (labday 2 - 5)
- May (labday 6 - 17)
- June (labday 18 - 45)
- July (labday 46 - 75)
- August part 1 (labday 76 - 92)
- August part 2 (labday 93 - 106)
- September part 1 (labday 107 - 123)
- September part 2 (labday 124 - 135)
- October part 1 (labday 136 - 149 )
- October part 2 (labday 150 - 166 )
- November (labday 167 - 170 )
- Cellculture
18. Labortag 01.06.2010:
Modifying MCS of pAAV_MCS vector
Investigators: Anissa, Adrian, Bea, Chris W., Hanna, Patrick, Volker, Sven
Oligos received from Sigma-Aldrich
(right ITR of pAAV_MCS, left ITR of pAAV_MCS and MCS RFC25 for pAAV)
- Hybrization of received oligos: MCS RFC25 for pAAV (forward) and MCS RFC25 for pAAV (reverse)
- Centrifuge tubes prior to open tubes (13.000 rpm, 30 sec)
- MCS RFC25 for pAAV (forward): Add 92µL Millipore H2O (Volume on obtained sheet)
- MCS RFC25 for pAAV (reverse): Add 394 µL Millipore H2O (Volume on obtained sheet)
- Vortex the resuspended DNA
- Make aliquots of both Oligos (1:10): 10 µL Oligo + 90 µLH2O (final volume usually 100 µl)
- Mix together(into PCR-tube):
| Volume/µL | solution |
| 10 (1:10) | Oligo 1: MCS RFC25 for pAAV (forward) |
| 10 | Oligo 2: MCS RFC25 for pAAV (reverse) |
| 4 | 100mM TrisHCl pH8 |
| 8 | 5mM MgCl2 |
| 8 | H20 |
- Program: ORIGAMI 1 modified for long oligos:
- 1 99°C 7’
- 2 99°C 1’
- -1°C R=0.3 °/s
- Goto 2 rep 74
- Hold 4°C
- While hybridization of oligos is performed, digestion of pAAV_MCS vector can be conducted
following standard protocol for cloning.
- Title: Ligation MCS_Oligo with pAAV_MCS
- Plasmid: pAAV_MCS
- Buffer used: 3
- BSA: Yes
- Measure DNA-concentration with Nanodrop
- DNA-Concentration:260 ng/uL
- Restriction-enzyms used: http://www.neb.com/nebecomm/DoubleDigestCalculator.asp
Enzyme1 (Nr. Lab: 152): ClaI
Enzyme2 (Nr. Lab: 15): BglII
- Digestion components :
| components | pAAV_MCS |
| DNA | 4 |
| BSA (10x) | 3 |
| Buffer 3 (10x) | 3 |
| Enzyme: ClaI (no.Lab:152) | 2 |
| Enzyme: BglII (no.Lab:15) | 1 |
| H2O | 17 |
| Total volume | 30 |
- Incubate for 1,5 h at 37°C
- 1% Agarose gel
- 1% agarose gel was prepared, gel ran for 45 minutes( first: 90V, after 15 minutes: 115 V)
- Amount of loading dye added
| sample/µL | loading dye/µL |
| marker: 8 | contains loading dye |
| pAAV_MCS: 24 | 6 |
- Expected size of fragments
| sample | expected size |
| pAAV_MCS: cut with ClaI and BglII | 4580 bp |
19. Labortag 02.06.2010:
Picking clones of pAAV_RFC25MCS
Investigators: Adrian, Bea, Chris W., Hanna, Anissa
Practical work:
Control plate contained no clones. :)
4 colonies were picked and grown @ 37°C over night.
Design and ordering of Oligos (NotI)
Investigators: Adrian, Bea, Chris W., Hanna, Anissa
Oligos for site directed mutagenesis of the NotI restriction sites in pAAV_MCS (ITRs) were designed:
Media:Freiburg10 NotI ITR Oligos.pdf
Sponsoring work:
Sponsoring letter was adapted for Quiagen.
20. Labortag 03.06.2010:
pAAV_RFC25_MCS -> problem
Investigators: Anissa, Bea, Melanie, Christian L.
Comment: Continue with Mini-Prep and test digestion of pAAV_RFC25_MCS
Mini-Prep and test digestion have been performed:
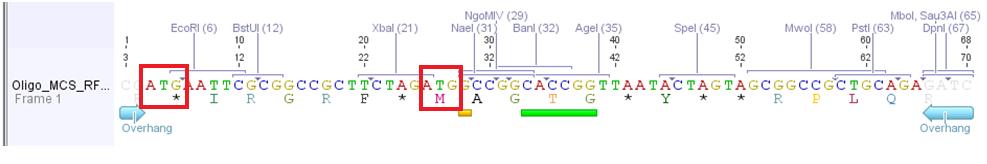
Problem: Designed oligos (MCS_RFC25) for altering the MCS of the pAAV_MCS vector cannot be used.
Two startcodons are in the MCS.
The two startcodons are not in the same open reading frame (ORF). Therefore two proteins will be produced. The Gene of Interest and the short peptide (30 aa).
The idea of the oligos was to ligate the oligos into the pAAV_MCS vector. The oligos contained two overhangs which correspond to the sequences of the two restriction sites ClaI and BglII:
The pAAV_MCS vector was digested with ClaI and BglII and then we ligated the oligo and the vector. Problem was that we did not notice that the overhang of ClaI and the sequence of EcoRI of the MCS_RFC25 resulted in another ATG startcodon.
Possible Solutions:
- first: modify MCS with ordered oligos of Sven (shorter MCS which cannot be used for pEX)and clone mVenus
- Perform site-directed-mutagenesis (QuikChange from Stratagene)
- Order new MCS-oligos and consider that no new ATG is produced. For example: add another base between ClaI overhang and EcoRI sequence. -----ATXG---- This solution is the more possible one we are going to perform.
Practical work
- Preparing four glycerol stocks (2:1)
- numbers: B4 - B7 (for details see nomenclature)
- stored in -80°C, Box 1
- MiniPrep
- Nanodrop concentrations
| Sample | Concentration/ng*µl-1 |
| P11 | 340,5 |
| P12 | 364,0 |
| P13 | 358,5 |
| P14 | 284,4 |
- Test digestion
| Components | Volume µl | Mastermix µl |
| DNA | 800 | -- |
| BSA (10x) | 1,5 | 7,5 |
| Buffer No.2 (10x) | 1,5 | 7,5 |
| Enzyme 1 (no.Lab:45) Nde I | 0,5 | 2,5 |
| Enzyme 2 (no.Lab:71) Spe I | 0,5 | 2,5 |
| H2O | variable | -- |
| Total volume | 15 | 20 |
| Sample | Volume/ µl | H2O / µl |
| P11 | 2,3 | 8,7 |
| P12 | 2,2 | 8,8 |
| P13 | 2,2 | 8,8 |
| P14 | 2,8 | 8,2 |
- Incubation: 1,5 h
- Materials
| Sample | Sample/µl] | Loading dye (5x/6x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| P11 | 15 µl | 3 µl | 3677 bp | 974 bp |
| P12 | 15 µl | 3 µl | 3677 bp | 974 bp |
| P13 | 15 µl | 4 µl | 3677 bp | 974 bp |
| P14 | 15 µl | 4 µl | 3677 bp | 974 bp |
| Marker | Sample P11 /18 µl | Sample P12 /18 µl | Sample P13 /19 µl | Sample P14 /19 µl | |
|---|---|---|---|---|---|
| Lane | 1 | 3 | 5 | 7 | 9 |
Results of agarose-gel:
- Expected fragments of 3677 bp and 974 bp can be verified. The insertion of the RFC25_MCS has been inserted.
Picking clones of Thymindinkinase of Amor
- 5 clones of the XL-1 Blue colonies containing the plasmid pUB6/HV5/His6 with the thymidinkinase-guanylate fusion enzyme have been picked from LBamp-agarplates
- 1 clone of the XL-1 Blue colonies containing the plasmid pUB6/HV5/His6 without the thymidinkinase (control) has been picked from LBamp-agarplates
- all clones have been inoculated in 10 mL LB containing 10 µL Amp. Incubation: 37°C over-night.
- to do: Mini-Prep of pUB6/HV5/His6 with the thymidinkinase and pUB6/HV5/His6 without the thymidinkinase (control)
Idea
- Insertion of Kozak consensus sequence before MCS to enhance gene expression (cloning consideration in Stratagene manual)
- New RFC standard with Kozak sequence for eucaryotes??
- pXX6 all Adenovirus helper genes without AAV genes
- pKEX-2XL.Rep40 expression plasmid for Rep protein 40
- pKEX-2XL.Rep 52 expression plasmid for Rep protein 52
- pKEX-2XL.Rep 68 expression plasmid for Rep protein 68
- pKEX-2XL.Rep 78 expression plasmid for Rep protein 78
- pCMV-VP(HS) expression plasmid for VP proteins in combination (assemly competent)
- pKEX-VP1 expression plasmid for single VP proteins (Cap: VP1, VP2 und VP3). Be careful: Single constructs are not assembly competent; Ruffing et al 1992; Steinbach et al. 1997;
- pKEX-VP2expression plasmid for single VP proteins (Cap: VP1, VP2 und VP3). Be careful: Single constructs are not assembly competent; Ruffing et al 1992; Steinbach et al. 1997;
- pKEX-VP3expression plasmid for single VP proteins (Cap: VP1, VP2 und VP3). Be careful: Single constructs are not assembly competent; Ruffing et al 1992; Steinbach et al. 1997;
- pTRUF_CMV_eGFPsingle stranded vector for eGFP expression
- dsAAV_CMV_eGFP"self complementary" expression vector from X. Xiao
- pTAV2 (whole AAV genome in "blue script" vector)
- pDG complete helper plasmid, nedded for vector production ; Grimm et al. 1998
- In order to obtain the DNA following steps have been performed:
- cut out the spot where the DNA is spotted with a clean scalpel (note: scalpel should not have any contamination).
- put a 0,5 mL Eppi in a 1,5 mL Eppi. Put little holes in the smaller eppi.
- transfer Whatman paper into Eppi
- add 50 µL TE-EF (Redissolving buffer) to whatman paper and wait 15 minutes
- centrifuge eppis at 2000 rpm, 10 minutes
- Transformation with obtained plasmids was performed.
21. Labortag 04.06.2010:
DKFZ plasmid Retrafos, TK/GMK Mini-Prep
Investigators: Adrian, Bea, Chris W., Hanna
DKFZ
Comments: Plasmids of PD Kleinschmidt of the DKFZ arrived. The DNA was dried on a whatman paper.
Plasmids received:
Fusion Enzyme: Thymidinkinase/Guanylate Kinase (TK/GMK)
Plasmid Mini-Prep
- experiment date: 04.06.2010 ; time: 3,5h
- name of investigator: Adrian, Bea, ChrisW.,Hanna
- new vector name: pUB_V5_His6 + TK/GMK (fusionenzyme)
Glycerol Stocks
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | Clone 5 | Control | |
| Bacteria strain | XL-1 Blue | XL-1 Blue | XL-1 Blue | XL-1 Blue | XL-1 Blue | XL-1 Blue |
| Plasmidname | pUB_V5_His6 + TK/GMK | pUB_V5_His6 + TK/GMK | pUB_V5_His6 + TK/GMK | pUB_V5_His6 + TK/GMK | pUB_V5_His6 + TK/GMK | pUB_V5_His6 + TK/GMK |
| Date | 04.06.2010 | 04.06.2010 | 04.06.2010 | 04.06.2010 | 04.06.2010 | 04.06.2010 |
| given number | B8 | B9 | B10 | B11 | B12 | B13 |
Given Plasmid-Number
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | Clone 5 | Control | |
| given number | P15 | P16 | P17 | P18 | P19 | P20 |
Nanodrop concentration
- Plasmid
- Given Plasmid-Number: P15; DNA concentration: 493,7 ng/µL ;
- Given Plasmid-Number: P16; DNA concentration: 464,4 ng/µL;
- Given Plasmid-Number: P17; DNA concentration: 445,6 ng/µL;
- Given Plasmid-Number: P18; DNA concentration: 562,9 ng/µL;
- Given Plasmid-Number: P19; DNA concentration: 528,1 ng/µL;
- Given Plasmid-Number: P20; DNA concentration: 499,9 ng/µL;
Comments:A Plasmid-Mini Prep with the received Fusionenzyme Thymidinkinase/Guanylate Kinase (TK/GMK)from Amor has been performed.
The DNA will be sent to GATC for sequencing.
- pUB6_V5_His6 - clone 1 (P15) (3 µL added to 27µL H20) has been sent to GATC.
- Expected results: Saturday
22. Labortag 05.06.2010:
Insertion of iGEM expression parts into pAAV_MCS
Investigators: Adrian, Bea, Melanie, Hanna
Hybridization:
- Hybridization of received oligos: iGEM expression parts (RFC25 without EcoRI, NotI)
- Prior to opening the tubes, they were centrifugated at 13.000 rpm for 30 sec.
- expression part MCS_for (charge-no: ST00114065): 108 µL Millipore H2O (Volume on obtained sheet)were added.
- expression part MCS_for (charge-no: ST00114066): 165 µL Millipore H2O (Volume on obtained sheet)were added.
- Resuspended DNA was vortexted.
- Aliquots of both Oligos (1:10) were prepared: 10 µL Oligo + 90 µL H2O (final volume usually 100 µl).
- Mix together(into PCR-tube):
| Volume/µL | solution |
| 10 (1:10) | Oligo 1: expression part MCS_for (charge-no: ST00114065) |
| 10 (1:10) | Oligo 2: expression part MCS_for (charge-no: ST00114066) |
| 4 | 100 mM TrisHCl pH8 |
| 8 | 5 mM MgCl2 |
| 8 | H20 |
- Program: ORIGAMI 1 modified for long oligos:
- 1 99°C 7’
- 2 99°C 1’
- -1°C R=0.3 °/s
- Goto 2 rep 74
- Hold 4°C
- While hybridization of oligos was performed, digestion of pAAV_MCS vector was conducted
following standard protocol for cloning.
Digestion:
- Title: Ligation iGEM expression parts (="iGEM-MCS") with pAAV_MCS
- Plasmid: pAAV_MCS
- Buffer used: 3
- BSA: Yes
- DNA-Concentration: 260 ng/uL
- Restriction-enzyms used:
Enzyme1 (Nr. Lab: 152): ClaI
Enzyme2 (Nr. Lab: 15): BglII
- Digestion components :
| components | pAAV_MCS |
| DNA | 5.8 µL |
| BSA (10x) | 3 µL |
| Buffer 3 (10x) | 2 µL |
| Enzyme: ClaI (no.Lab:152) | 2 µL |
| Enzyme: BglII (no.Lab:15) | 1 µL |
| H2O | 16.2 µL |
| Total volume | 30 µL |
- Mixture was incubated for 1,5 h at 37°C.
Agarose-Gel:
- 1% agarose gel was prepared, gel ran for 45 minutes(110 V)
- Amount of loading dye added
| sample/µL | loading dye/µL |
| marker: 8 | contains loading dye |
| pAAV_MCS: 30 | 6 (6x loading dye) |
- Expected size of fragments
| sample | expected size |
| pAAV_MCS: cut with ClaI and BglII | 4580 bp |
</ul>
Gelextraction:
Gel measurement:
| Sample | weight |
| pAAV_MCS | 60 mg |
- Gelextraction was performed following standard protocol.
- DNA-concentrations were meassured: pAAV_MCS 18.2 ng/µL, Oligos: 136.7 ng/µL -> 1:10 dilution was prepared: 13.67 ng/µL
Ligation:
| iGEM-MCS | pAAV_MCS | |
| Volume/µl | 0.4 | 8.6 |
Trafo was performed (using XL1B cells) following standard protocol.
23. Labortag 07.06.2010:
Mini-Preps (Kleinschmidt-plasmids and pAAV_iGEM-MCS)
Investigators: Investigators: Achim, Kira,Jessy, Chris W., Hanna, Adrian, Bea
pAAV_iGEM-MCS
Plasmid Mini-Prep
- experiment date:07.06.2010; time: whole day
- name of investigator: Achim, Kira,Jessy, Chris W., Hanna, Adrian, Bea
Glycerol Stocks
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | |
| Bacteria strain | XL1B | XL1B | XL1B | XL1B |
| Plasmidname | pAAV_iGEM-MCS | pAAV_iGEM-MCS | pAAV_iGEM-MCS | pAAV_iGEM-MCS |
| Date | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 |
| given number | - | B27 | B28 | B29 |
Given Plasmid-Number
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | |
| given number | - | P34.2 | P34.3 | P34.4 |
Test digestion
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 (no. Lab:___) AgeI ; Enzyme 2 (no.Lab:___) NdeI
- Plasmid
- Given Plasmid-Number: P34.2; DNA concentration: 433.78 ng/µL ;
- Given Plasmid-Number: P34.3; DNA concentration: 408.48 ng/µL ;
- Given Plasmid-Number: P34.4; DNA concentration: 409.80 ng/µL ;
Comments:Clone no.1 was discarded.
Test digestion:
| Components | Volume/µL | Mastermix |
| DNA (clone 2) | 1000 ng | - |
| BSA (10x) | no | - |
| Buffer no. 4 (10x) | 1.5 µL | - |
| Enzyme 1 (no. Lab: ) AgeI | 0.75 µL | - |
| Enzyme 2 (no. Lab: ) NdeI | 0.5 µL | - |
| H2O | variable | - |
| Total volume | 15 µL | - |
| Sample | Volume sample/ µl | Volume H2O / µl |
| P34.2 | 2.3 | 9.95 |
| P34.3 | 2.4 | 9.85 |
| P34.4 | 2.4 | 9.85 |
- Incubation: 45 min, 37°C
Agarose-Gel:
0.5 g Agarose, 50 ml TAE (1%), 3 µL GELRED (3-6µl), at 110 Volt, running time: 45 minutes
| Sample | Sample/µl] | Loading dye (5x/6x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| P34.2 | 15 µl | 3 µl | 3686 bp | 951 bp |
| P34.3 | 15 µl | 3 µl | 3686 bp | 951 bp |
| P34.4 | 15 µl | 3 µl | 3686 bp | 951 bp |
- Marker: GeneRuler ladder mix
| Marker | Sample | Sample | Sample | |
|---|---|---|---|---|
| Lane | 34.2 / 18 µl | 34.3 / 18 µl | 34.4 / 18 µl |
Kleinschmidt-plasmids
Investigators: Kira, Achim, Jessy, Bea, Adrian, Hanna
Plasmid Mini-Prep
- experiment date: 07.06.2010 ; time: 10 – 20 h
- Kleinschmidt-plasmids
Glycerol Stocks
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | Clone 5 | Clone 6 | Clone 7 | Clone 8 | Clone 9 | |
| Bacteria strain | XL1B | XL1B | XL1B | XL1B | XL1B | XL1B | XL1B | XL1B | XL1B |
| Plasmidname | pXX6 | pKEX-2XL.Rep 40 | pKEX-2XL.Rep 52 | pKEX-2XL.Rep 68 | pKEX-2XL.Rep 78 | pCMV-VP(HS) | pKEX-VP1 | pKEX-VP2 | pKEX-VP3 |
| Date | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 |
| given number | B14 | B15 | B16 | B17 | B18 | B19 | B20 | B21 | B22 |
| Clone 10 | Clone 11 | Clone 12 | Clone 13 | |
| Bacteria strain | XL1B | XL1B | XL1B | XL1B |
| Plasmidname | pTRUF_CMV_eGFP | dsAAV_CMV_eGFP | pTAV2 | pDG |
| Date | 07.06.2010 | 07.06.2010 | 07.06.2010 | 07.06.2010 |
| given number | B23 | B24 | B25 | B26 |
Given Plasmid-Number
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | Clone 5 | Clone 6 | Clone 7 | Clone 8 | Clone 9 | Clone 10 | Clone 11 | Clone 12 | Clone 13 | |
| given number | P21 | P22 | P23 | P24 | P25 | P26 | P27 | P28 | P29 | P30 | P31 | P32 | P33 |
| measured concentration | 351,01 | 673,1 | 532,22 | 579,05 | 725,31 | 659,68 | 692,8 | 545,46 | 568,34 | 420,62 | 446,95 | 496,8 | 472,58 |
Comments:
Many things went wrong today!
- Glycerol stocks must be vortexted!
- Check mini-prep buffers - especially buffer PE (needs to contain ethanol!)
- Always check volumes - try to estimate if volume makes sense (check pipettes!)
- Don't discard bacteria cultures until glycerol stocks and mini-preps are successfully done!!!
Today's conclusion: Better ask 2 times than do something wrong without asking!!!
Site-directed mutagenesis of pAAV_iGEM-MCS (PstI)
Quickchange site directed mutagenesis:
PCR reaction:
- 2.5 µL 10x Pfu Ultra II buffer
- 0.5 µL template (therefore the a 1:20 dilution of the pAAV_iGEM-MCS (433 ng/µL) was prepared) = 10.825 ng
- 0.56 µL primer 1 (of 1:10 dilution)
- 0.56 µL primer 2 (of 1:10 dilution)
- 1 µL DMSO (primers form very strong secondary structures)
- 0.5 µL dNTP
- 18.88 µL dH2O
- 0.5 µL PfuUltra II fusion (1.25 U)
-> end volume: 25 µL
PCR program:
1 x : 2' 95°C (HotStart polymerase)
20 x : 30 s 95°C -> 1' 55°C -> 5' 68°C
1 x : 4°C (over night)
Experiment will be continued tomorrow.
24. Labortag 08.06.2010:
Cloning of mVenus_YFP into pAAV_iGEM-MCS, continuation of site-directed mutagenesis
Investigators: Kira, Jessy, Hanna, Achim
Analysis of iGEM-MCS sequence (RFC25 without EcoRI and NotI):
The alignment with the theoretical pAAV_iGEM-MS delivered a "C-deletion" within the NgoMIV restriction site. Due to that the pAAV_iGEM-MCS lacks this site. We controlled the bill of delivery and noted that the deletion must be GATC's fault.
All in all this doesn't matter, because parts which are in iGEM standard can be inserted and therefore they will deliver the lacking NgoMIV restriction site. Further on this could be an advantage because after digestion the fragment which is cut out can be better detected in the gel.
Insertion of mVenus_YFP into pAAV_iGEM-MCS
Digestion:
| components | V (pAAV_iGEM-MCS)/ µl | I(pGA14mVenus_Geneart) / µl |
| DNA | 2,3 | 3,8 |
| BSA (10x) | 2 | 2 |
| Buffer 3 (10x) | 2 | 2 |
| Enzyme: AgeI (no.Lab:149) | 1,25 | 1,25 |
| Enzyme: XbaI (no.Lab:63) | 0,75 | 0,75 |
| H2O | 11,7 | 10,2 |
| Total volume | 20 | 20 |
0.5 g Agarose, 50 ml TAE (1%), 3 µL GELRED (3-6µl), at 115 Volt, running time:
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| P34.2 | 20 µl | 4 µl | 4617 bp | 22 bp |
| pGA14mVenus | 20 µl | 4 µl | 2870 bp | 774 bp |
| Sample | weight | concentration |
| Vektor_Oben | 0,32 g | 29,8 ng/µl |
| Vektor_Unten | 0,14 g | 12,8 ng/µl |
| Insert | 0,3 g | 12,9 ng/µl |
- Vector: 4,16 µl
- Insert: 4,84 µl
- Vector: 6 µl
- Insert: 3 µl
Continuation of site-directed mutagenesis
Digestion with DpnI:
Trafo:
25. Labortag 09.06.2010:
pAAV_iGEM-MCS_mVenus-YFP, Site-directed mutagenesis
Investigators: Jessy, Achim, Sven, Toby, Hanna
No pAAV_iGEM-MCS_w/oPstI transformed bacteria (site-directed mutagensis) grew on the ampicillin agar plates over night!
Experiment is conducted again, guided by Toby (Thanks a lot!!!).
Further on the cloning of mVenus-YFP into pAAV_iGEM-MCS seemed to fail:
Only on the agar plate cotaining the bacteria transformed with the "vector_oben" ligation, which actually should be the "wrong" ligation (gelextraction of a band which contained fragments that were too large, see picture in lab journal), revealed colonies. Therefore this experiments is also conducted one more time, guided by Sven (thanks also a lot!!!)
Insertion of mVenus_YFP into pAAV_iGEM-MCS
- experiment date: 9.6.2010
- plasmid:
- Vector: name: pAAV_iGEM-MCS number: 34.2 production date: 5.6.2010
- Insert: name: pOG14_mVenus number: - production date: ____ origin: Sven :)
- new vector name: pAAV_iGEM-MCS_mVenus-YFP
- buffer used: NEB4 ; Restriction-enzymes used: Enzyme 1 (no. Lab:___) AgeI ; Enzyme 2 (no.Lab:___) XbaI
- DNA concentration (vector): 433 ng/µL ; DNA concentration (insert): 530 ng/µL
Digestion
| components | volume of vector /µl | volume of insert /µl |
| DNA | 2.83 | 2.77 |
| BSA (100x) | 0.5 | 0.5 |
| Buffer NEB4 (10x) | 3.0 | 3.0 |
| Enzyme 1 (AgeI) | 1.75 | 1.75 |
| Enzyme 2 (XbaI) | 1.25 | 1.25 |
| H2O | 20.73 | 20.67 |
| Total volume (e.g. 15,20,25,30 µl) | 30 | 30 |
Agarose-Gel:
0.5 g Agarose, 50 ml TAE (1 %), 3 µL GELRED (3-6µl), at 115 Volt
Gelextraction
- insert (mVenus-YFP): 26 ng/µL
- vector (pAAV_iGEM-MCS): 30 ng/µL
Ligation
- vector: 5.85 µL
- insert: 3.15 µL
- ligase: 1 µL
- buffer: 10 µL
Test digestion of pAAV_iGEM-MCS
p style="font-size:15px; font-weight: bold; color: blue;">Test digestion</p>
- buffer used: 1 ; Restriction-enzymes used: Enzyme 1 (no. Lab:___) NgoMIV
- Plasmid
- Given Plasmid-Number: P34.2 ; DNA concentration: 433.78 ng/µL;
- Given Plasmid-Number: P34.3 ; DNA concentration: 408.48 ng/µL;
- Given Plasmid-Number: P34.4 ; DNA concentration: 409.8 ng/µL;
| Components | P34.2 [µL] | P34.3 [µL] | P34.4 [µL] |
| DNA | 2.3 | 2.5 | 2.4 |
| BSA (10x) | 1 | 1 | 1 |
| Buffer no. 1 (10x) | 1 | 1 | 1 |
| Enzyme 1 (no. Lab: 113) ngoMIV | 1 | 1 | 1 |
| H2O | 4.7 | 4.5 | 4.6 |
| Total volume | 10 | 10 | 10 |
- Incubation: 1 h, 37°C
Agarose-Gel:
0.5 g Agarose, 50 ml TAE (1 %), 3 µL GELRED, at 115 Volt, running time: 45 minutes + 20 minutes
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| P34.2 | 10 µl | 2 µl | 3728 bp | 903 bp |
| P34.3 | 10 µl | 2 µl | 3782 bp | 903 bp |
| P34.4 | 10 µl | 2 µl | 3782 bp | 903 bp |
- Marker: GeneRuler ladder mix
| Marker | Sample P34.2 /µl | Sample P34.3 /µl | Sample P34.4 /µl | |
|---|---|---|---|---|
| Lane | 8 µL | 10 µL | 10 µL | 10 µL |
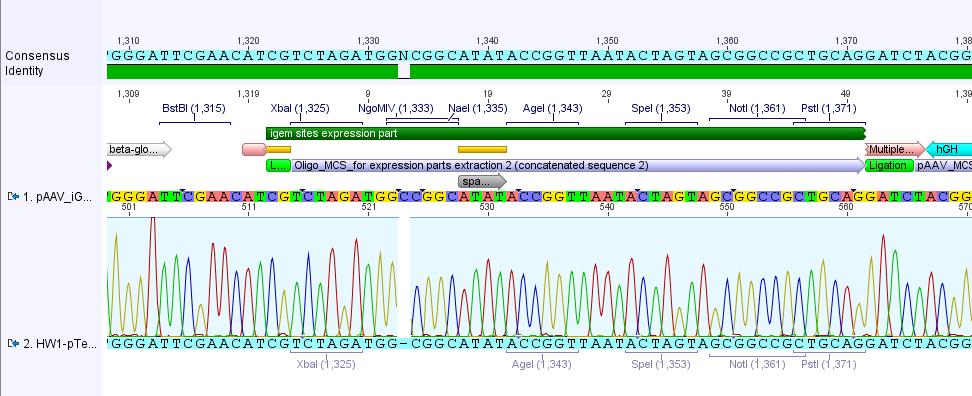
Comments: Digestion of P34.2 delivered just one fragment size, revealing that - as expected after sequencing (C-deletion: no NgoMIV site in iGEM-MCS) - there was just one NgoMIV restriction site in the vector. In contrast to that the digestion of P34.3 and P34.4 delivered two bands - whereas the band of the smaller fragments looked like a smier. Because of this obscure and non-satisfying results (besides our assumption that there's something wrong with the marker :) )we decided to sequence the P34.3 vector:
- c(P34.3): 408.48 ng/µL
- Volume (plasmid): 5.14 µL
- Volume (vector): 24.86 µL
- Name of eppi: HW_34
- Primer: GATC_std_pTeSp-1
Picking of Clones from pAAV_iGEM-MCS_mVenus-YFP_Vektor_oben
Investigator: Hanna, Achim
- experiment date: 9.6.2010
- plasmid:
- Vector: name: pAAV_iGEM-MCS_Vektor_oben
- Insert: name: pOG14_mVenus
- new vector name: pAAV_iGEM-MCS_mVenus-YFP_Vektor_oben
The Clones were picked and incubated in 10 mL LB-Medium containing 10µL ampicillin over night at 37°C on a rotary shaker.
26. Labortag 10.06.2010:
Site-directed mutagenesis of PstI in ITRs
1. Site directed mutagenisis
- Aim: deletion of PstI from both ITR's
- 2 PCR tubes got prepared, one for the left ITR and one for the right. (The SDM was performed according to the standart protocol)
- Media:Freiburg10_Site_directed_Mutagenesis_pAAV_MCS_deletion_PstI.pdf
Mini-Prep of pAAV_iGEM_Venus_YFP and test digestion
Investigators: Jessy, Achim
2.1 Mini-Prep of pAAV_iGEM_mVenus_YFP
Title: pAAV_iGEM_mVenus_
Glycerol Stocks
| Clone 1 | Clone 2 | |
| Bacteria strain | XL1 blue | XL1 blue |
| Plasmidname | pAAV_iGEM_mVenus | pAAV_iGEM_mVenus |
| Date | 10.06.2010 | 10.06.2010 |
| given number | B30 | B31 |
Given Plasmid-Number
| Clone 1 | Clone 2 | |
| given number | P35 | P36 |
Title: pAAV_iGEM_mVenus_YFP (guided by Sven)
investigator: Bea
Glycerol Stocks
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | |
| Bacteria strain | BL-21 | BL-21 | BL-21 | BL-21 |
| Plasmidname | pAAV_iGEM_mVenus_YFP | pAAV_iGEM_mVenus_YFP | pAAV_iGEM_mVenus_YFP | pAAV_iGEM_mVenus_YFP |
| Date | 10.06.2010 | 10.06.2010 | 10.06.2010 | 10.06.2010 |
| given number | B32 | B33 | B34 | B35 |
Given Plasmid-Number
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | |
| given number | P37 | P38 | P39 | P40 |
2.2 Test digestion
- buffer used: 4 ; Restriction-enzymes used: Enzyme XbaI (no. Lab:___) ; Enzyme AgeI (no.Lab:___) ____
- Plasmid
- Given Plasmid-Number: P37; DNA concentration: 114,8 ng/µL ;
- Given Plasmid-Number: P38 ; DNA concentration: 179,8 ng/µL;
- Given Plasmid-Number: P39 ; DNA concentration: 180,8ng/µL ;
- Given Plasmid-Number: P40; DNA concentration: 189,2 ng/µL;
Comments: Clones were picked from trafo-plate in the morning and mini-prep was performed in the late afternoon. Due to this and the fact that bacterial strain was BL-21, DNA-concentrations are quite low.
Investigators: Bea, Chris W.
| Components | Volume/µL | Mastermix | Sample: P37 | Sample: P38 | Sample: P39 | Sample: P40 |
| DNA | 800 ng | - | 7,0 µL | 4,5 µL | 4,4 µL | 4,2 µL |
| BSA (10x) yes | 1,5 µL | 7,5 µL | MM 4,5 µL | MM 4,5 µL | MM 4,5 µL | MM 4,5 µL |
| Buffer no. 4 (10x) | 1.5 µL | 7,5 µL | " | " | " | " |
| Enzyme 1 (no. Lab: ) AgeI | 0.75 µL | 3,75 µL | " | " | " | " |
| Enzyme 2 (no. Lab: ) XbaI | 0.5 µL | 2,5 µL | " | " | " | " |
| H2O | variable | - | 3,75 µL | 0,25 µL | 6,35 µL | 6,55 µL |
| Total volume | 15 µL | - | 15 µL | 15 µL | 15 µL | 15 µL |
- Incubation: 45 min, 37°C
27. Labortag 11.06.2010:
Site directed mutagenisis of 10.06.2010
Transformation
Transformation was performed according to the standard protocol Media:Freiburg10_Cloning Protocol.pdf
Cells were plated on agar plates (2x 1:100; 2x pellet)containing ampicillin and stored over night at 37°C.
Sequencing
p39 was send to GATC for sequenzing
- p39: 180,83 ng*µl^-1
- volume plasmid: 8,3 µl
- volume water: 21,7 µl
- primer: GATC_std_pTeSp-1
28. Labortag 12.06.2010:
Transformation plates stored in 4°C room
Investigator: Kira
The plates were stored in the iGEM shelf @ 4 °C. Even if the transformation was not very successfull, some colonies can be picked and inoculated either tomorrow or on Monday.
Note: Site-directed mutagenesis does not generate lots of intact plasmids, so few colonies are normal!! (Tobias)
29. Labortag 13.06.2010:
Investigators: Jessy, Patrick, Hanna
We weren't able to detect any colonies on the plates - just a few air bubbles :) To be entirely sure, we put them back into the 37°C room over night.
30. Labortag 14.06.2010:
pAAV_iGEM-mVenus-YFP:
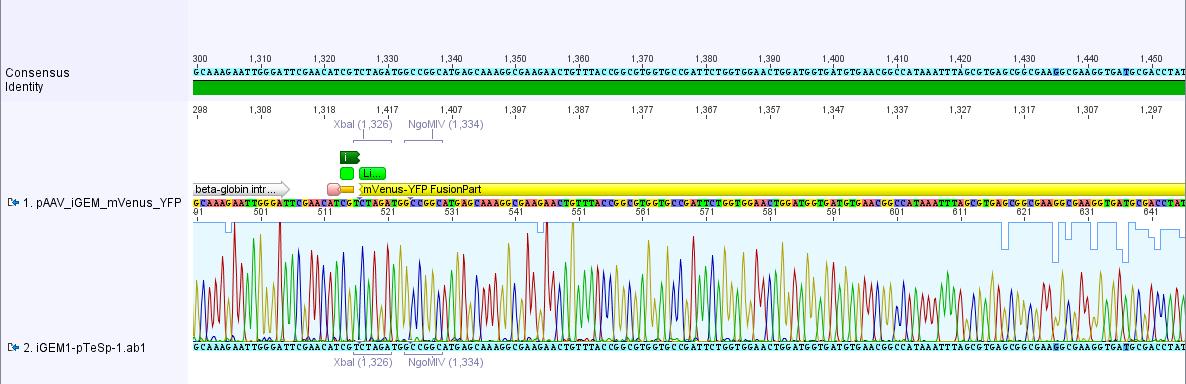
The sequencing data of pAAV_iGEM_mVenus-YFP was analyzed. The alignment with the "theoretical" pAAV_iGEM_mVenus-YFP showed that we successfully inserted mVenus-YFP into pAAV_iGEM-MCS.
Site-directed mutagenesis:
Also today no colonies were detectable on any plates!
TK-GMK-Plasmid
Investigators: Adrian, Hanna
puB6_V5_His6_clone1 + TK/GMK (P15) will be sequenced again:
- name: AF 1
- primer: GATC_std_BGH-reverse
- volume(plasmid) = 4.25 µL
- volume (H2O) = 25.75 µL
Results (15.06.): Unfortunately just ~ 300 bp were sequenced.
To do: Therefore more primers have to be designed in order to sequence the whole ~2000 bp TK-GMK fusion construct!
Cell culture
AAV 293 and HT1080 Cells have been splitted and plated out on 10 cm dishes for transfektion. The Cells were accounted by using the Neubauer-Meteringchamber. After harvesting by following the standard protocol the pallet have been resuspendiated in 15ml of DTT medium. 2,5µl of this cell suspension have been mixed with 47.5µl of trypan blue.
We counted 12,5x 10^6 cells/ml for the T293 AAV cell line and 10x10^6 cells /ml for the HT1080 cell line.
The AAV 293 cells have been plated out on four dishes with 250µl of the cell suspension, on one plated with 1ml and on another dish with 1.5ml. note that that the date is wrong on the plates and the flasks! Cells have been already plated out on the 14th. of June.
We also seated two flasks one for each cell line. Therefor we used 1ml of the HT1080 cell suspension and two ml of the the AAV293 cells.
31. Labortag 15.06.2010:
Solutions for transfection were prepared
Investigators: Adrian, Achim, Chris W., Hanna
- 1 x TE-Buffer: 1.2114 g TRIS, 0.2 mL EDTA, 81 mL milipore-H2O were mixed. pH was adjusted with HCl (1M and 5M) to 7.50. Volume was adjusted to 100 mL with milipore-H2O. Solution was autoclaved (sterilization by filtration is also possible).
- 1 M CaCl2: 147.02 g CaCl2 x 2H2O (M = 147.02 g/mol) was solved in 1 Liter H2O and sterilized by filtration.
- 2 x HBS: 0.027 g Na2HPO4, 1.636 g NaCl, 1.19155 g HEPES were dissolved in ~ 30 mL milipore-H2O. pH was adjusted to 7.10. Volume was adjusted to 100 mL and sterilized by filtration.
32. Labortag 16.06.2010:
Sponsoring:
Investigators: Anna, Kerstin, Anissa
Sponsoring letters were examined and modified one more time.
Sequencing of TK-GMK:
Investigator: Hanna
Two primers were designed and ordered from Sigma-Aldrich. Probably we will receive them on Friday.
Mega primer PCR:
Investigators: Bea, Hanna
33. Labortag 17.06.2010:
Primer designing: VP1 primer for pKEX forward/reverse
Investigator: Volker
The construnts that we receieved from PD Dr. Kleinschmidt (DKFZ) are mainly in the pKEX backbone. The sequences of the backbone and the inserts are not availible, for this reason we decided to sequence the constructs. The first step is to sequence from one construct into the backbone. In the second step we will designe primers that bind in the backbone and point into the direction of the insert.
These primers will then be used to sequence the inserts of all the constructs.
Forward primer pKEX in VP1 from bp 4124 to 414
- VP1 primer for pKEX forward: 5' - CAACAAGTCTGTTAATGTGGAC - 3' 22bp; TM: 50°C; CG% 41%
Reverse primer pKEX in VP1 from bp 2081 to 2100
- VP1 primer for pKEX reverse: 5' - GTGGGCCAGGTTTGAGCTTC - 3' 20bp; TM: 54°C CG%: 60%
The amplicon that should be produced by the primers is 199 bp long.
Primer designing: CMV_forward/reverse_qPCR
Investigator: Volker
Primers for the titering of the CMV region from the literature [Rohr et al., 2002] and [Rohr et al., 2005] were compared and analysed.
The sequences from 2002 showed a differce in one nucleotide compared to the other primer and to our sequence. For this reason the second set [Rohr et al., 2005] was ordered.
- CMV_forward_qPCR: 5' - GGGACTTTCCTACTTGGCA - 3'
- CMV_reverse_qPCR: 5' - GGCGGAGTTGTTACGACA - 3'
These primers will be used in a quantitative PCR assay with Sybr-green to measure the genomic titer and the infectious titer of the viral particles we will produce in the future.
Ordering of required reagents:
Investigator: Volker
- Nuclease S1 was ordered from Promega (10000Units for 36,00€)
- Proteinase K was ordered from Sigma (5mg for 31,30€; BioUltra, ≥30 units/mg protein, lyophilized powder)
These reagents are required for the procedure to determine the genomic and the infectious titer.
34. Labortag 18.06.2010
Transfection
Transfection via calcium phosphat with the prepared solutions (31. Labortag 15.06.2010)
Investigators: Chris W., Hanna, Patrick, Adrian, Volker
Transfections with the following plasmids:
1)
- pAAV_iGEM_mVenus_YFP (glycerol stock B34, P39) P39 has the correct sequence (confirmed)
- pHelper
- pAAV_RC
2)
- pAAV_iGEM_mVenus_YFP (glycerol stock B33, P38) P38 (we dont know if P39 has the correct sequence!)was used because the amount of P39 was not sufficient enough for four Transfections!
- pHelper
- pAAV_RC
Plasmid concentrations:
pAAV_RC: 1 µg/µl
pHelper: 280 ng/µl
pAAV_iGEM_mVenus_YFP, P39: 180,83 ng/µl
pAAV_iGEM_mVenus_YFP, P38: 179,75 ng/µl
Cellculture clone 1 and 2 were transfected with P39. Cellculture clone 3 and 4 were transfected with P38.
Deviations from the standard protocol (currently incomplete): We could only pipet 3,3 µg (instead of 10 µg) from each of the 3 plasmids into the 15 ml falcons due to insufficient amount of plasmid.
Adrian tried to examine the precipation of the CaCl2+ DNA Clusters. We have to optimize the pH of our 2xHBS at the moment it is 11.12
Theoretical: Digestion of pAAV_MCS for PCR to design ITR BioBrick
Investigator: Bea
Idea:
- Digest vector in order to obtain two fragments which contain the left and the right ITR respectively.
- Separation (Agarose-gel) and gel extraction of fragments
- Perform PCR with iGEM-Primers (forward primer contains EcoRI and Xbai; reverse primer contains SpeI and PstI)
- to do:
- design primers
- digestion of pAAV_MCS etc.
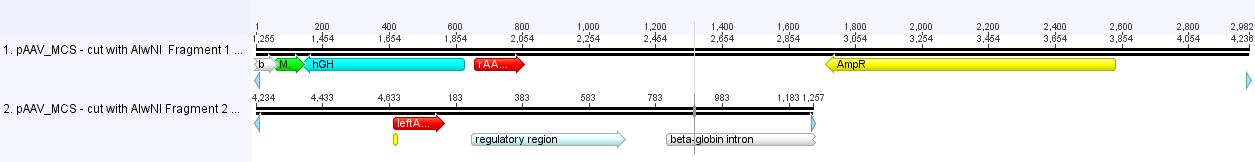
Theoretical: Digestion of pAAV_MCS with AlwNI produces two fragments which can be separated by agaorse gel.
35. Labortag 19.06.2010: Literature seminar
Investigators: Bea, Hanna, Melanie, Denis, Volker, Christian W., Adrian and Achim
Topics:
- The ITRs
- The Cap
36.Labortag 20.06.2010:
Picking clones of sequenced pAAV_igEM_mVenus_YFP
Investigators: Bea and Chris W
- Incoluate 250 mL DYT medium (add 250µL ampiciliin) with B34 (glycerol stock: BL-21 E.coli cells with P39 plasmid: pAAV-iGEM_mVenus_YFP
- Incubate on rotary shaker over-night (o/n) at 37°C
- to do: perform Midi-Prep tomorrow (21.06.2010)
37.Labortag 21.06.2010:
Second Transfection, Transduction
Investigators: Patrick, Johannes
2x HBS was steril filtrated into two 50 ml flacons. The falcons were put into the cellculture fridge.
New medium is prepared:
DMEM-Medium
- 500ml DMEM with Glutamax
- 10% FCS (50 ml)
- 5 ml 100x Pen/Strep
- 5 ml 100x NaPyruvat
H-DMEM-Medium
- 400 ml DMEM with Glutamax
- 90 ml FCS
- 5 ml 100x Pen/Strep
- 5 ml 100x NaPyruvat
Feching dry ice for the experiments
Investigators:Volker
Dry ice can be fetched from the department for macromolecular chemistry. The chemical depot is located in the second basement. The dry ice can be retrieved from monday to friday from 11:00 to 11:30 and from 14:15 to 14:30. The requestforms for this institute can be found in the iGEM folder and should be filled out, signed by an advisor and taken to the chemical depot. A styrofoam container is required for the transport.
Annotation of the BRs and the PLA2 domains
Investigator:Volker
The Bacis Regions which are important for the nuclear localisation were annotated in the protein sequences of VP 1-3 according to:
[Grieger et al, 2006]
Endotoxin-free Midi-Prep of pAAV_iGEM_mVenus-YFP
Investigators: Chris W., Hanna (guided by Sven)
Midi-Prep was performed following standard protocol: File:Freiburg10 Endotoxinfreie Midi.pdf
DNA concenration was measured: 507 ng/µL
Plasmid was further used for a...
Second Transfection
Investigators: Patrick, Johannes
Six transfections were performed according to the standart protocol: Media:Freiburg10_Transfection_protocoll.pdf
Three 10 cm cellcluture dishes with 293 cells were transfected with 10 µg DNA (3,33 µg each plasmid) and the other three 10 cm cellculture dishes with 30 µg DNA (10 µg DNA each plasmid).
Plasmids:
- pAAV_iGEM-MCS_mVenus (P41), 507 ng/µl
- pAAV_RC , 1000 ng/µl
- pHelper , 280 ng/µl
Preparing Viral Stocks
Investigators: Patrick, Johannes
According to the standard protocol (see AAV Helper-free System manual). No dilution was performed.
HT1080 Transduction
Investigators: Patrick, Johannes Four 10 cm cellculture dishes with HT1080 cells have been provided for the transduction.
- 1 ml of the AAV-2-mVenus (derived from P 39) was pipeted to into dish 1. Note: the virus solution (buffer) showed a yellow coloration.
- 1 ml of the AAV-2-mVenus (derived from P 39) was pipeted to into dish 2. Note: the virus solution showed (buffer) an orange coloration.
- 1 ml of the AAV-2-mVenus (derived from P 38) was pipeted to into dish 3. Note: the virus solution showed (buffer) an orange coloration.
- 1 ml of the AAV-2-mVenus (derived from P 38) was pipeted to into dish 4. Note: the virus solution showed (buffer) an orange coloration.
Apart from the dilution, the transduction was performed according to the standart protocol.
38.Labortag 22.06.2010:
Midiprep of pHelper plasmid; cloning of the rfc25 MCS
Investigators: Bea, Chris W, Adrian, Achim
- Hybridisation of oligos RFC25 EcoR1+BglII for & RFC25 EcoR1+BglII rev
- Programm ORIGAMI1 was modified for normal oligos:
- 1 95°C 7'
- 2 95°C 1'
- -1°C R=0,3°s
- Rep 70
- Programm ORIGAMI1 was modified for normal oligos:
- conc of Oligos: 138,3µg/µl
Digestion
| components | volume of vector /µl | volume of insert /µl |
| DNA | 5,8 | none |
| BSA (100x) | none | none |
| Buffer 3 (10x) | 3 | none |
| Enzyme EcoRI (no.Lab:23) | 1,5 | none |
| Enzyme BglII (no.Lab:15) | 1,5 | none |
| H2O | 18,2 | none |
| Total volume (e.g. 15,20,25,30 µl) | 30 | none |
- A 1% agarose gel was prepared for plasmid separation
- Production of antibiotika-stock-solutions (10 x 1ml AMP)
- Midi Prep of pAAV-Helper: concentration. 732,6 µg/µl
- dilution of our Oligos RFC from 138,3 µg/µl 1:10 13,83 (71bp long)=> 0,64µl
- Vektor pAAV-MCS 23.3 µg/µl => 8,36µl
- two plates are placed in the 37°C room
- Trafo has been performed following standard protocols
39.Labortag 23.06.2010:
Preparation of Viral Stock, Seeding cells
Design of Primer
Investigators: Achim, Hanna
In order to sequence the hGH (polyadenylation) sequence and in general the GOI (gene of interest :) ) primers were designed: Media:Primer GOI+polyA pAAV iGEM-MCS mVenus YFP.pdf
These primer will be used in order to sequence P38 (pAAV_iGEM_mVenus-YFP) which delivered fluorescencing HT1080 cells - in comparison to the already sequenced P39 clone. Further on they can be used in general for sequencing any GOI.
These oligos were ordered at sigma aldrich as E@sy Oligos.
P38 was send for sequencing with GATC_std_pTESp-1 primer: 12 µL P39 (180 ng/µL) + 18 µL H2O
Sequencing of pKEX-VP1
Investigators: Achim, Hanna, Adrian, Chris W.
Primer:
- VP1 primer for pKEX forward: V = 3 µL + 27 µL H2O
- VP1 primer for pKEX reverse: V = 3 µL + 27 µL H2O
Vector:
- pKEX-VP1 (P27.2): c = 693 ng/µL, V = 3 µL + 27 µL H2O
Thaw HT1080 and AAV-293 cells
Investigators: Achim, Hanna (guided by Sven)
Thawing of HT1080 and AAV-293 cells was performed following this protocol: Media:Freiburg10 Thawing cells.pdf
Picking clones of pAAV_RFC25
Investigators: Achim, Chris W., Hanna
Clones were picked, 250 mL DYT were inoculated.
Bacteria cultures are stored at 37°C over night.
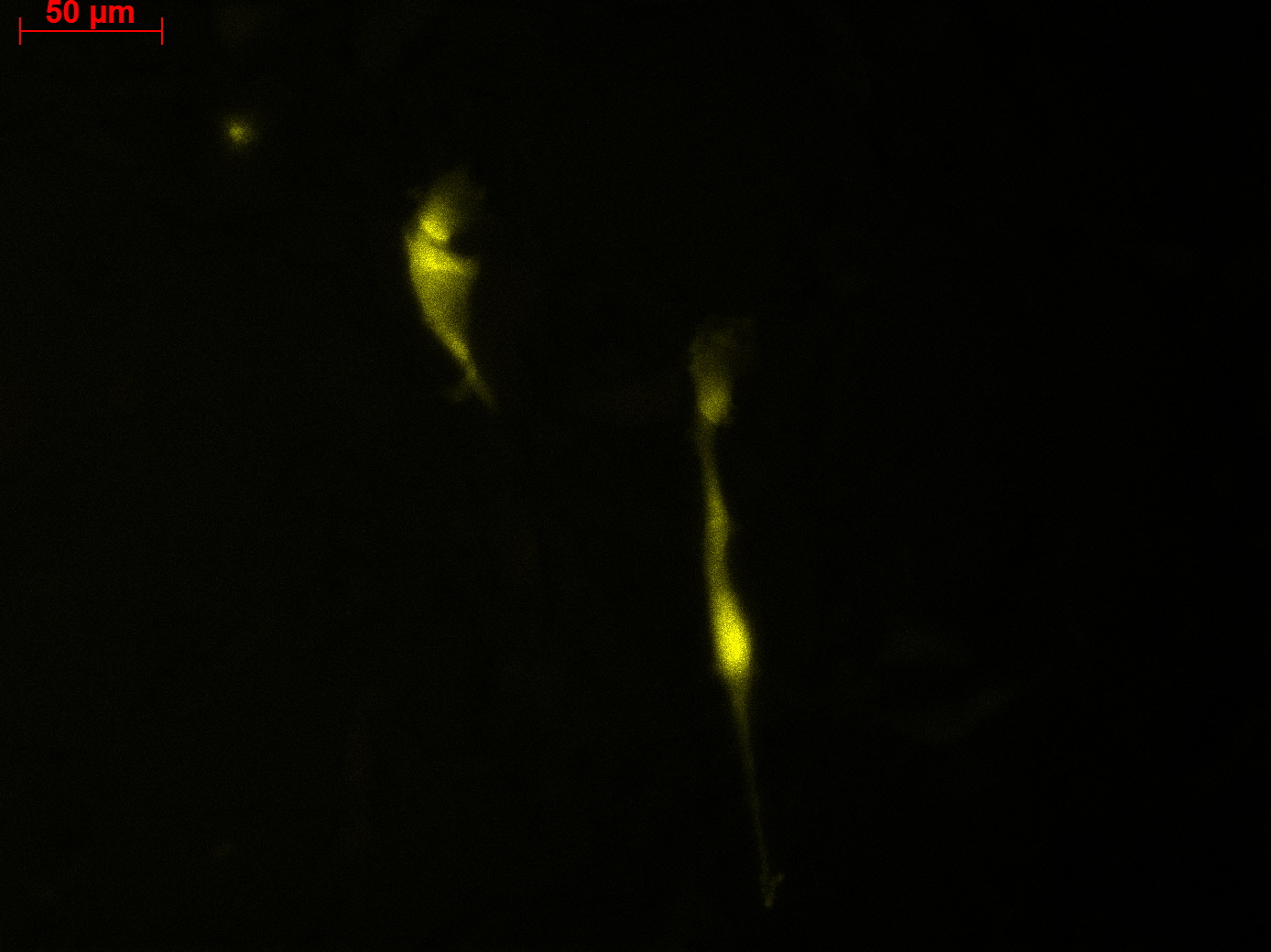
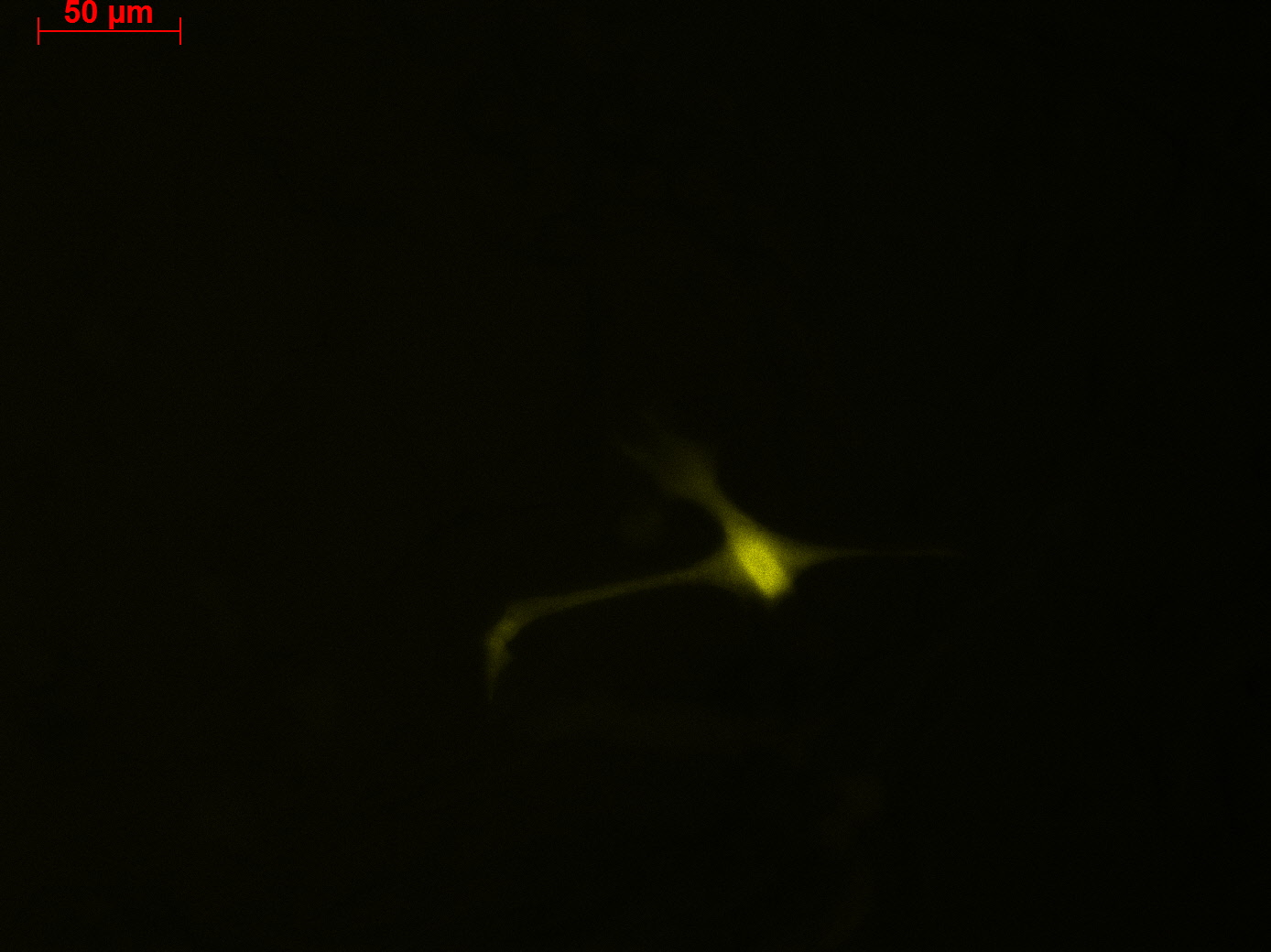
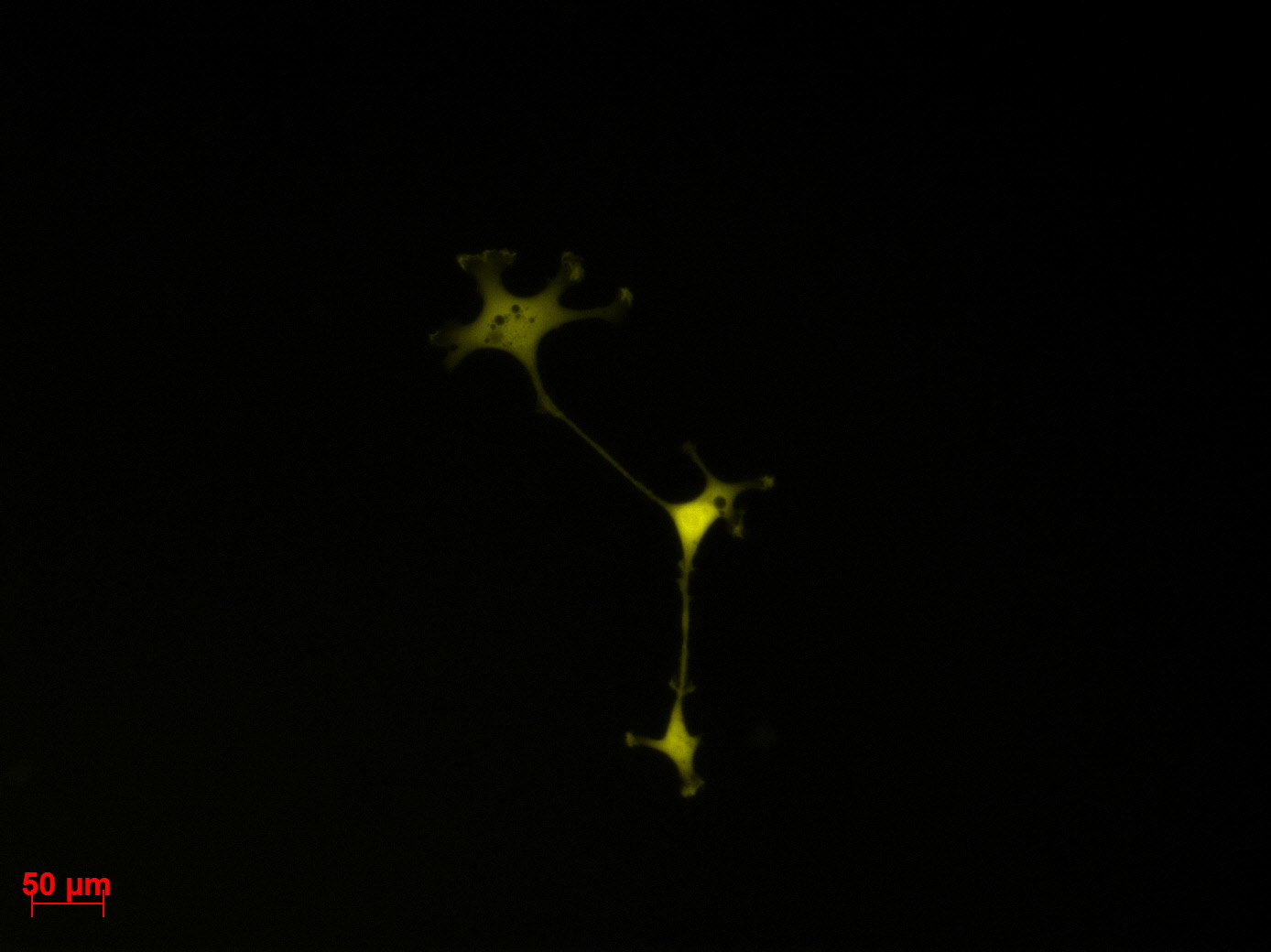
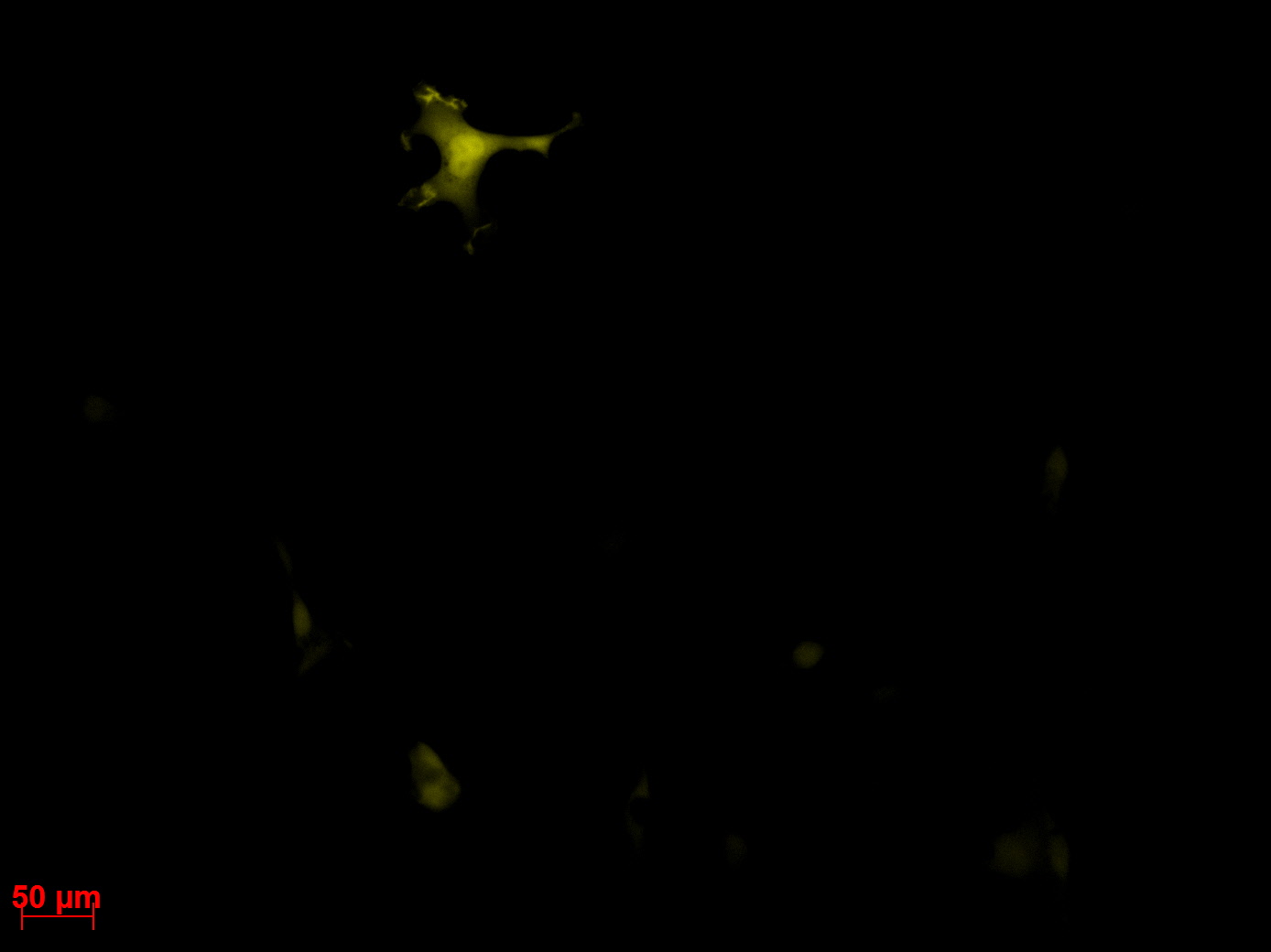
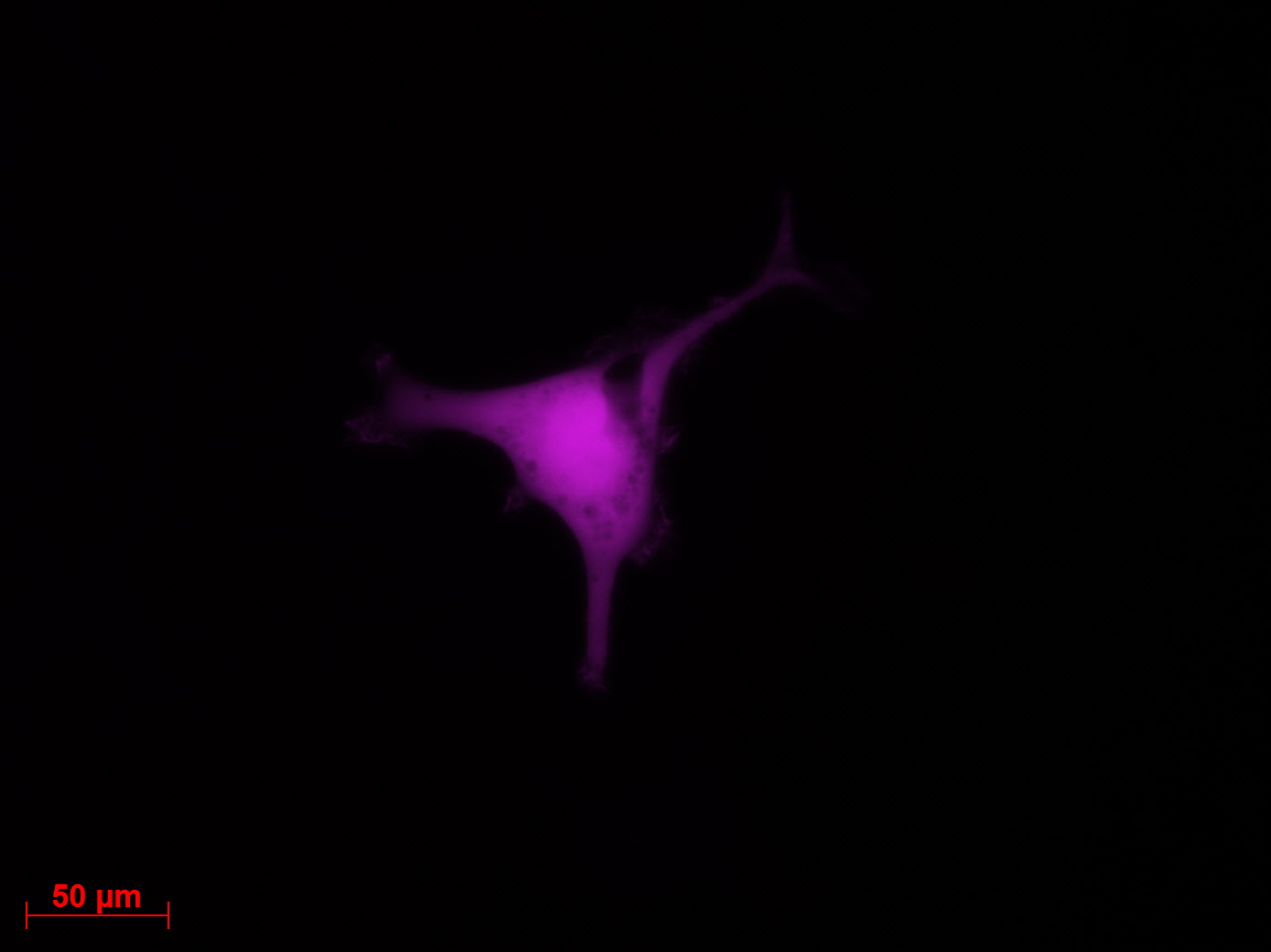
Transduction results
Investigators: Adrian (guided by Sven)
Preparation of viral stocks
Investigator: Johannes, Bea
Steril filtration of virus particles was performed (guided by Sven). Viral stocks are stored at -80°C.
40.Labortag 24.06.2010:
Labmeeting, Presentation/Update by Patrick and Adrian
Delivery of:
Proteinkinase K (will be used for the degradation of the viral capsid prior to qPCR)
Benzonase (will be used fur viral stock preparation)
Primer for for ITR-conversion to iGEM-standars
- Presentations of Adrian and Patrick: Media:Adrian.ppt Media:Patrick.ppt
Midi-Prep of pAAV_iGEM_mVenus-YFP
Investigators: Hanna, Chris W (Adrian inserted RNAse and Ethanol into Buffers)
- new kit from Qiagen was used
- 35 ml of the over-night culture was filled into a 50 mL falcon and centrifuged at 5000 g at 4°C for 5 minutes.
- supernatant was discarded
- Midi-prep was performed according to manual of Qiagen
- final concentrations: 380,4 µg/µl
- Name of eppi: P44
Mini Prep of P42 and P43 pAAV-RFC-25
Investigator: Bea
- Mini-Prep was performed following standard protocols
- conc: P42: 321,5 µg/µl
- conc: P43: 310,1 µg/µl
ITR modification via PCR
- Primers for ITR modification were resuspended and labeled: left forward: o24, left reverse: o25, right forward: o26, right reverse: o27
Work on pKEX backbones
Investigator: Volker
The sequencing data was evaluated and BLAST searches were carried out on NCBI to answer the question weather the sequence belongs to the backbone pKEX. This question could be answered positively.
There are standard Primers from GATC that prime in the sequence. The three expression plasmids from PD Kleinschmidt will therefor be sequenced tomorrow.
Additionnaly nine primers for sequences in the AAV Rep and Cap genes were designed.
41.Labortag 25.06.2010:
PCR of ITRs, Splitting HT1080 and AAV-293 cells, Seeding of HT1080 cells, Creating Virus Stock Economics, Medium check
Preparation for PCR of ITRs
Investigator: Bea
Digestion of pAAV_MCS (P9)
- experiment date: 25.06.2010
- plasmid: name: pAAV_MCS number: P9 production date: 08.05.2010 origin: SH
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 (no. Lab:113) NgoMIV ; Enzyme 2 (no.Lab:120) AlwNI
- DNA concentration (vector): 267,9 ng/µL
In order to receive the best PCR-results, two digestion reactions/approaches have been performed.
The first digestion was cut only with AlwNI, the second digestion was cut with AlwNI and NgomIV.
| components | v(pAAV_MCS 1) /µl | v(pAAV_MCS 2) /µl |
| DNA | 4 | 4 |
| BSA (100x) | none | none |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme AlwNI (no.Lab: 120) | 1 | 1 |
| Enzyme NgoMIV (no.Lab:113) | none | 1 |
| H2O | 13,0 | 12,0 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 |
Splitting of HT1080 and AAV-293 cells, Seeding of HT1080 cells
Investigator: Hanna
HT1080:
- Cells showed 80 % confluence
- Cells were washed with PBS, detached with Trypsine
- 10 mL DMEM was added, cell suspension was centrifuged for 5 minutes at 200 g.
- Supernatant was discarded, pellet was resuspended with 8 mL DMEM.
- Cells were counted: 47.5 µL Trypan blue (stains dead cells) and 2.5 µL cell suspension were mixed and applied onto a Neubauer counting chamber -> 2.2 x 20 x 10^-4 cells per mL
- Therefore 27 µL of the cellsuspension was added to each 3 mL DMEM in the 6-well plates. -> 2 6-well plates were prepared for transduction!
- Further on cells were splitted: 110 µL cell suspension was pipetted into 2 75T flasks each containing 20 mL DMEM and stored in the 37°C incubator.
AAV-293 cells:
- Cells showed ~ 50% confluence
- Cells were splitted following the standard protocol.
Creating Virus Stock Economics
Investigator: Adrian
Medium check
Investigator: Adrian
- three little culture bottles got prepared (H-DMEM, DMEM and PBS) and set into the incubator. In the following days they'll get checked for bacerial growth.
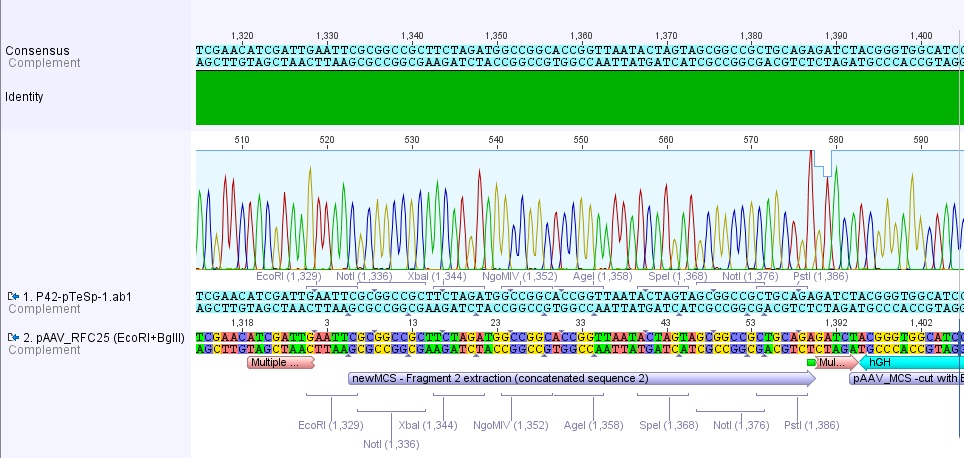
Sequencing results of pAAV_RFC25
Investigators: Adrian, Bea, Hanna
sequenced sample: pAAV_RFC25 clone 2 (P43)
used primer: GATC_std_pTeSp-1.ab1
The alignment with the theoretical construct showed, that there's a exchange of one base in the RFC25-multiple cloning site. This substitution leads to the deletion of the SpeI restriction site.
Therefore we checked the order sheet (everything OK!).
We sent the second clone (P42) for sequencing (Hanna).
- V(Plasmid) = 6.5 µL
- V(H2O) = 23.5 µL
- Primer: pTESP-1
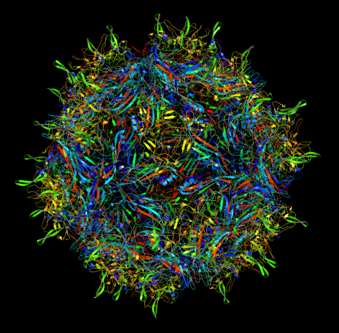
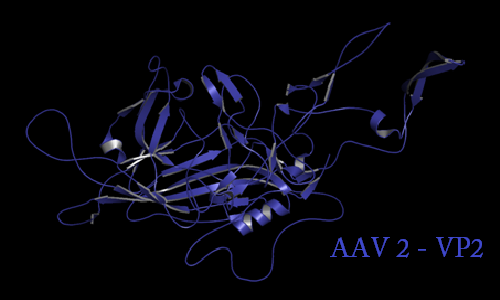
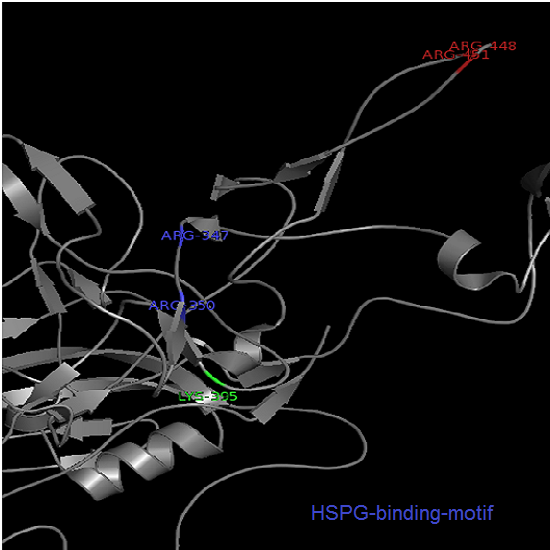
Annotation and studies of the AAV structure
Investigator: Volker
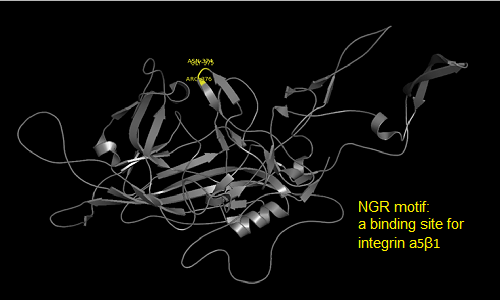
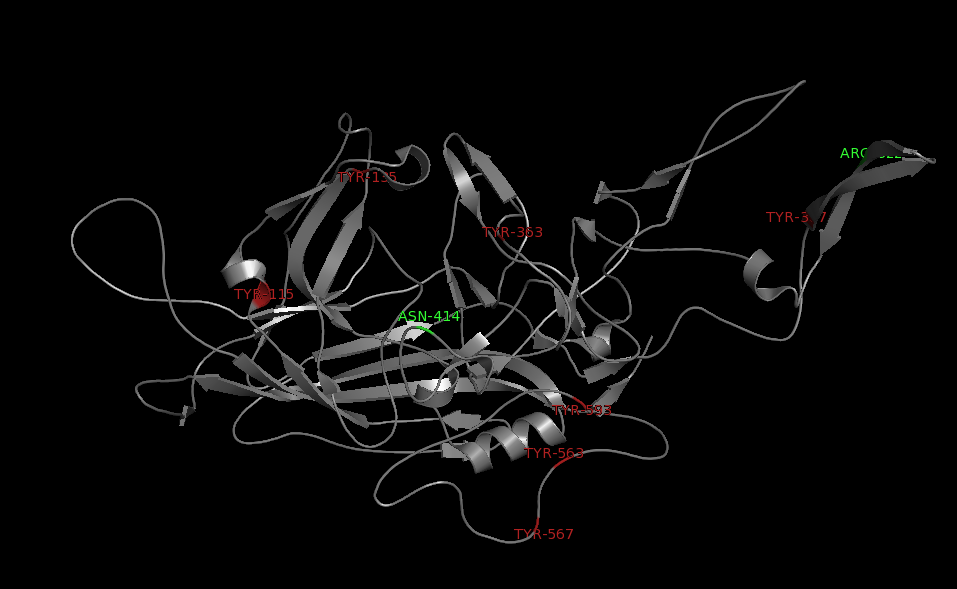
The NGR- and the HPSG motif were annotated in the gene sequences according to [Michelfelder & Trepe; 2009] The pdb sequence were visualized with the programm [http://www.pymol.org| PyMOL] and the important motifs were highlighted.
Impressions from PyMOL
42.Labortag 26.06.2010
Transduction of HT1080 cells
Investigator: Adrian
- Transduction of 2x6 wells was successfully done (14.25)
- one six well is transduced with 10µg the other with 30µg DNA amount (see HEK293 infection protokoll for further information)
(A is up)
| stock solution | 1:10 Dilution | 1:50 Dilution |
| 1:100 Dilution | 1:500 Dilution | control (500µl DMEM) |
- the delution steps (1:10, 1:50, 1:100, 1:500) are placed upon the virus stocks in the -80°C freezer in 15 ml falcons.
Did primers for sequencing GOI arrive?
Paper reading session
Investigators: Igor, Anna, Patrick, Stefan and Volker
Several papers were read and infomation on cap-modifications were extracted.
- Trasitions of seven Tyrosines (252, 272, 444, 500, 700, 704 and 730) to Phenylalanines as described in [Zhong et al. 2008]
- Transitions of two amino acids (R459D and N551D) leading to reduced sero prevalence as described in [Inchan Kwonand, David V. Schaffer]
43.Labortag 28.06.2010:
Checking HT1080 for YFP-Expression, Creating plan: urgent things to do
Investigator: Sven, Patrick, Adrian
Checking HT1080 for YFP-Expression
- Hannas 2*6 wells (24.6) transduced by (Adrian 25.6)
- The transduced cells will be checked again (29.6.).
Sequencing results of P42
Investigator: Hanna
Sequencing of pAAV_RFC25 (P42):
Sequencing delivered, that there weren't any base exchanges or deletions in this clone. Therefore P43 was dismissed (Plasmid and glycerol stock).
Theoretical construction of our biobricks
Investigators: Hanna, Adrian, Chris W, Patrick, Volker Theoretical cloning
Urgent Things to do:
- Modification CAP:
- HSPG -> 2 AS
- Targeting (585,588...) -> 2 AS -> 1 or 2 restriction sites?! Investigator: Volker
- Seroprevalence -> 2 AS
- thyrosine for ubiquitination Y252F, Y272F, Y444F, Y500F, Y700F, Y704F and Y730F
- Primer: start codons
- EGF-Rezeptor: A431-Zellinie
- Affibody: order the Gensequence
- bispecific AK -> order Diabodys (are there any restriction sites?) Investigator: kristian???
- Ordering the ITRs Investigator: Hanna
- Stratagene: right and left
- wild-type
- Modification: REP Investigator: Volker
- 3 restriction sites -> site directed mutagenisis vs. synthetic? ->order (check which is the cheaper approach)
- HGH Investigator: Hanna
- restriction sites? -> are there any available biobricks?
- biobrick production
- Beta-globin: what is the function? -> cut out?
- where is the start and the end?
- restriction sites?
- biobrick production
- GMK-TK
- restriction sites: primer
- biobrick production
- ordering the SR39
- tumor specific promotors
- more Information!!!
- telomerase specific promoters!!!
Continuation of ITR PCR
Investigators: Hanna, Chris W., Adrian
- Analytic gel was run: 1.7 µL loading dye (6x) was added to 8.3 µL of each sample (left ITR: 5 µL DMSO + 10 µL DMSO; right ITR: 5 µL DMSO + 10 µL DMSO).
- 110 V, 35 minutes
- No ITr bands were detectable - just 2 weak "primer-bands"
Conclusion: PCR didn't work. Because of that new primers were designed and ordered (Achim), which are longer and posses therefore higher annealing temperatures.
ITRs (theoretical)
Investigators:Adrian, Achim, Hanna
- trs sequences (GTTGG) are present in both ITRs (Stratagene)!
- assumption: transcription despite of ITR-secondary structure - no single-strand nick at trs-sequence in transduced cells ?
44.Labortag 29.06.2010:
Discussionround
- Affibodys
- Investigator: Anna
- Affibodys are 6-7kD Proteins derived from Protein A/Z
- they are binding to domain 3 from IGFR
- 13 AS are replaceable for changing the tropism
- Investigator: Anna
- Tyrosine Mutants
- Investigator: Adrian
- replacements are done in VP3! => Volker?!
- no multi-mutants are available (instead of single mutations many Y->F)
- best candidates are: Y730F and Y444F.
- Investigator: Adrian
- Second strand DNA-synthesis in transduced cells
- Investigator: Achim
- Second strand synthesis is the main limiting for transduction/transgen expression.
- Investigator: Achim
Seeding HT-cells for transfection and titering
Investigators: Sven, Adrian, Bea
- The pellet was resolved in 4ml we had a 550.000 cells/ml
- 2x6 well dishes got prepared:
1. Plate I(A is up)
| 3x10^5 cells | 3x10^5 cells | 3x10^5 cells |
| 3x10^5 cells | 3x10^5 cells | 3x10^5 cells |
2. Plate II (A is up)
| about 3x10^5 cells | only medium | only medium |
| about 3x10^5 cells | only medium | only medium |
Seeding HT1080-cells for transduction and qPCR
Investigator: Hanna
- Cell density was determined with trypan blue and counting chamber: 2 x 20 x 10^4 cells/mL
- wanted: 5 x 10^4 cells --> 125 µL cell suspension per well
- Cells were seeded into a 24-well-plate (row C and D!): 1.5 mL DMEM + 125 µL cell suspension per well
- cells were incubated for 1 hour at 37°C
Further on, cells were splitted: ~ 3 mL cell suspension was added to ~ 17 mL DMEM
Transduction of HT1080 cells for qPCR
Investigator: Chris W.
45.Labortag 30.06.2010:
Titering Procedure via qPCR, Transduction of 6well plates no.4
Harvesting cells for qPCR
Investigators: Chris W, Hanna
Transduced HT1080 cells were harvested following Sven's standard protocol.
Transduction of 2 x 6well plates nr.4
Investigators: Bea, Adrian
- Master plan: Different Transduction Mechanisms
- rise the amount of transduced particels, instead of 500µl, 1000µl of the AAV-stock getting pipetted into each well.
- the second plate gets stimulated via UV-light to boost the expression of DNA-Repair mechanisms. This should enhance the second strand synthesis of our ssDNA-GOI => a rise of YFP Expression!
1. Plate I(A is up)
| 3x10^5 cells | 3x10^5 cells | 3x10^5 cells |
| 3x10^5 cells | 3x10^5 cells | 3x10^5 cells |
Transduction:
| 1000 µl of Viral Stock DROPPED | 1000 µl of Viral Stock gently resuspended | 1500 µl of Viral Stock hard resuspending |
| 1000 µl of Viral Stock DROPPED | 1000 µl of Viral Stock gently resuspended | no Transduction |
2. Plate II (A is up)
| about 3x10^5 cells | only medium | only medium |
| about 3x10^5 cells | only medium | only medium |
Transduction:
| 1500 µl of Viral Stock gently resuspended | no Transduction | no Transduction |
| 1500 µl of Viral Stock gently resuspended | no Transduction | no Transduction |
beta-globin intron
Investigators: Adrian, Bea
We found some interesting literature which points out that introns have an influence on the eucaryotic gene expression on DNA and RNA level. Next steps are going to be the designing of the beta-globin intron and maybe further analysis of the other mentioned introns. Maybe two or three more introns can be ordered.
Media:Freiburg10_LeHir_et_al_How_introns_influence_and_enhance_eukaryotic_gene_expression_2003.pdf
Media:Freiburg10_Noe_et_al_An_intron_is_required_dro_dihydrofolate_reductase_protein_stability_2008.pdf
Media:Freiburg10_Mashadrizeh_et_al_A_systematic_study_of_the_function_of_the_human_beta-globin_introns_on_the_expression_of_the_human_coagulation_factor_in_cultured_Chinese_hamster_ovary_cells_2009.pdf
hGH sequence
Investigator: Hanna
- orientation in pAAV_MCS is reverse annotated, but in the same orientation as in human growth hormone I gene!
- Blast and alignment with other expression vectors and the human growth hormone I revealed that Stratagene annotated 1 bp too much
- "A common feature of mRNA in higher eukaryotes (but not in yeast) is the presence of the highly conserved sequence AAUAAA in the region from 11-30 nucleotides upstream of the site of poly(A) addition. (...) The signal is needed for both cleavage and polyadenylation." (Genes VIII, Lewin, P. 720)
- good news: There're no iGEM restriction sites in the sequence (RFC25)
- Conclusion: Instead of ordering the whole sequenc in iGEM standard it would be probably easier (and cheaper) to perform a PCR.
- Done: Oligos were designed for PCR!
ITRs: new strategy
Investigator: Hanna
Plan:
- Digest pAAV_MCS with AlwNI. Result: one large fragment containing the left ITR + a small fragment containing right ITR.
- Digest both fragments with NotI and PstI.
- Hybridize oligos containing RFC10 restriction sites (designed by Hanna), ligate them via intermediate steps (ligating into a vector) with ITRs . Comment: One oligo posseses a blunt end - therefore "intermediate step-vector" needs to be digested with an enzyme producing a blunt site.
- Done: Oligos were designed. Need to be checked!
Sequencing of pAAV_iGEM-mVenus-YFP
Investigators: Hanna, Chris W.
- 3 µL of O28, 29 and 30 each were mixed with 27 µL water -> primer fpr sequencing of GOI and of hGH sequence
- 11.1 µL plasmid + 18.9 µL H2O
Investigation of the Kleinschmidt sequencing
Investigator: Volker
The sequencing of den unknown backbone pKEX revealed that the construct VP1ex from the DKFZ seemed not to contain a regulatory region for the expresion of the VP1 ORF, but in the backbonesequence there were some standard primers from GATC that could be used to sequence the insert of all expression constructs.
This sequencing was done at the 26.06.2010. The read into the construct VP1 showed a long homologous region with the ORF of VP1 where as the sequencing of VP2ex and VP3ex did not show a homologous region with the ORFs of VP2 and VP3.
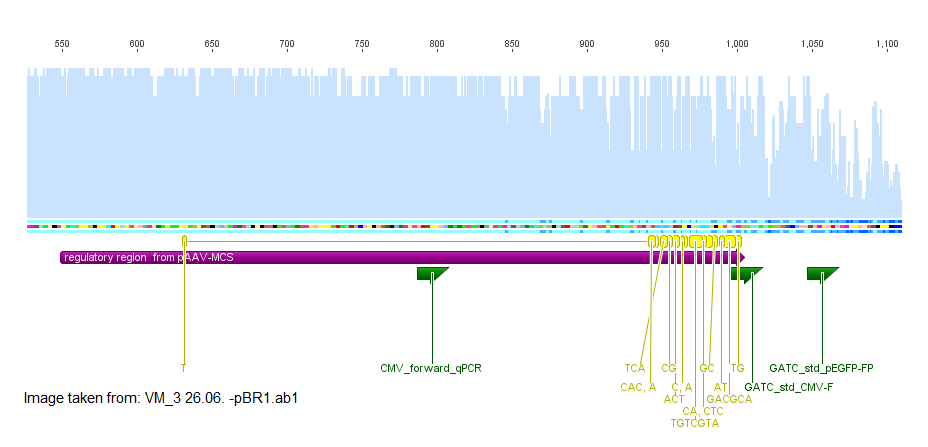
The test weather primers of GATC bind to the unknown region gave a positive hit in both constructs with the primer GATC_std_CMV_f. This primer is specific for the sequence of the CMV promoter. After this indice the sequences VM_2 26.06. -pBR1.ab1 and VM_3 26.06. -pBR1.ab1 were aligned with the sequence of the CMV promoter provieded in the stratagene kit in the plasmid pAAV-MCS. The majority of the sequence fitted well with CMV promoter as indicated in the image in purple. At the 5' end of the CMV promoter there were some missmatches in the alignment indicated in yellow.
The fact that VP2ex and VP3ex contained a CMV promoter element which was not included in VP1ex was unexpected. The sequencing with the standard primer GATC_std_CMV_f which binds at the 3' end of the CMV promoter and will probably sequence the ORF of VP2ex and VP3ex was ordered.
 "
"