Team:Heidelberg/Project/miRNA Kit
From 2010.igem.org
Laura Nadine (Talk | contribs) |
Laura Nadine (Talk | contribs) (→Results) |
||
| Line 41: | Line 41: | ||
==Results== | ==Results== | ||
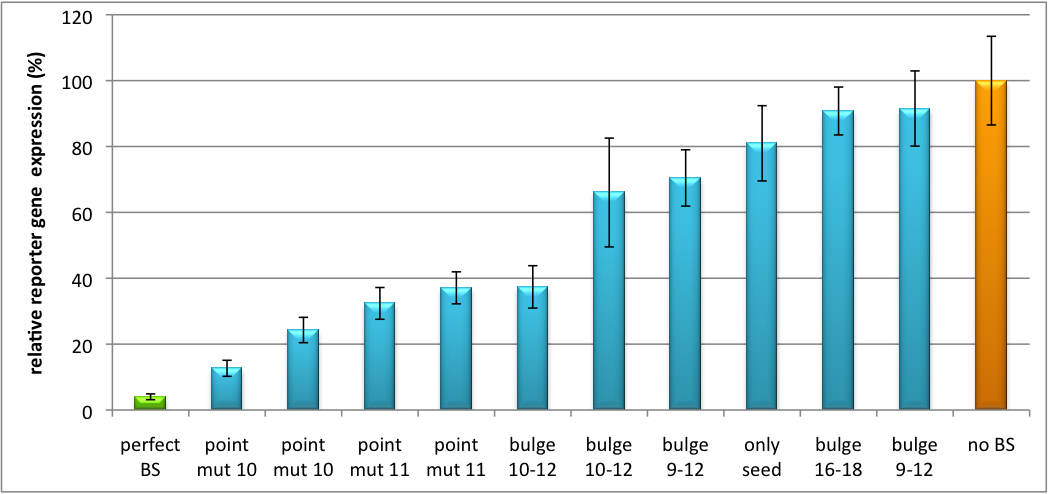
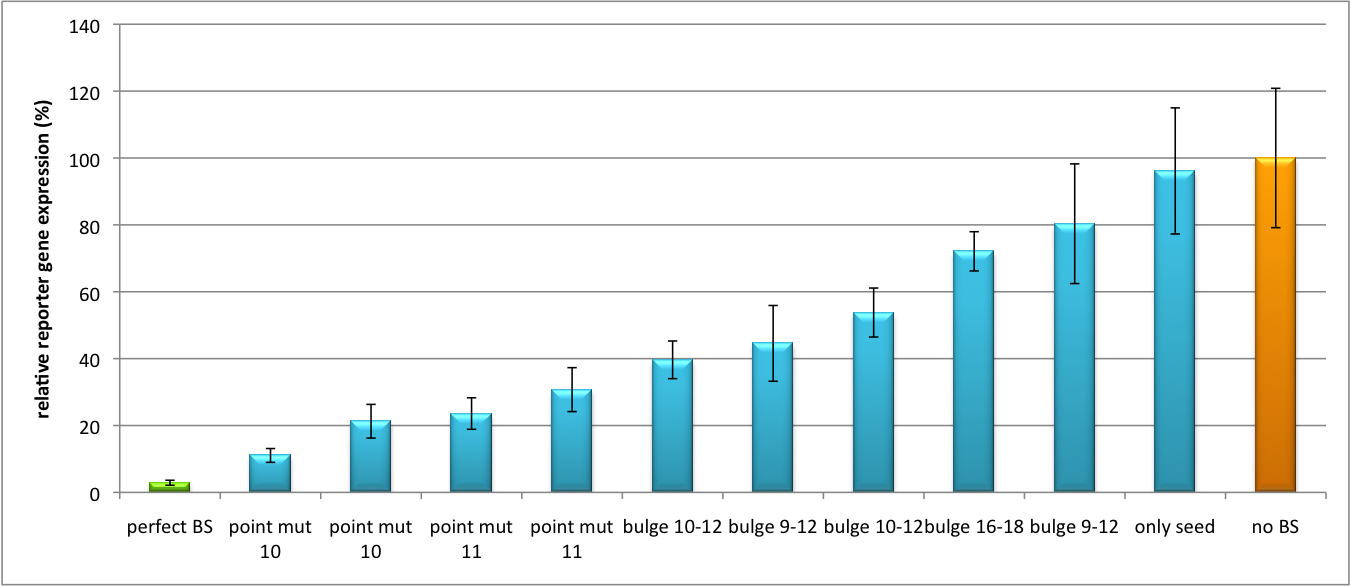
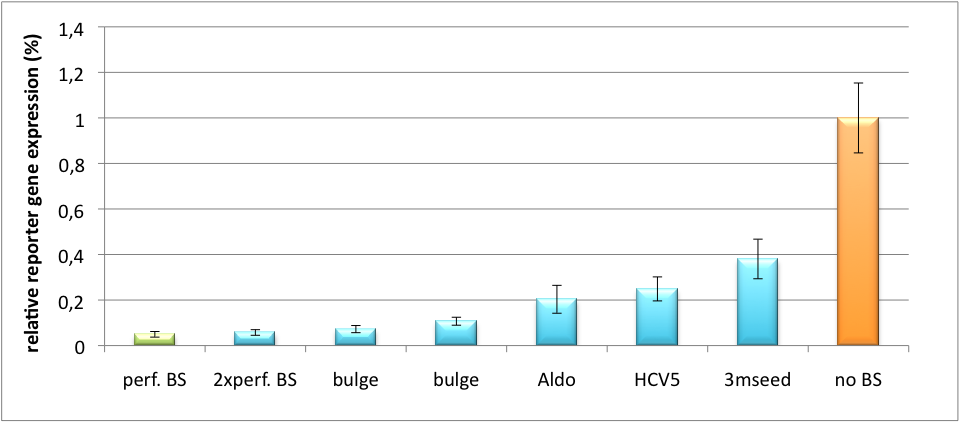
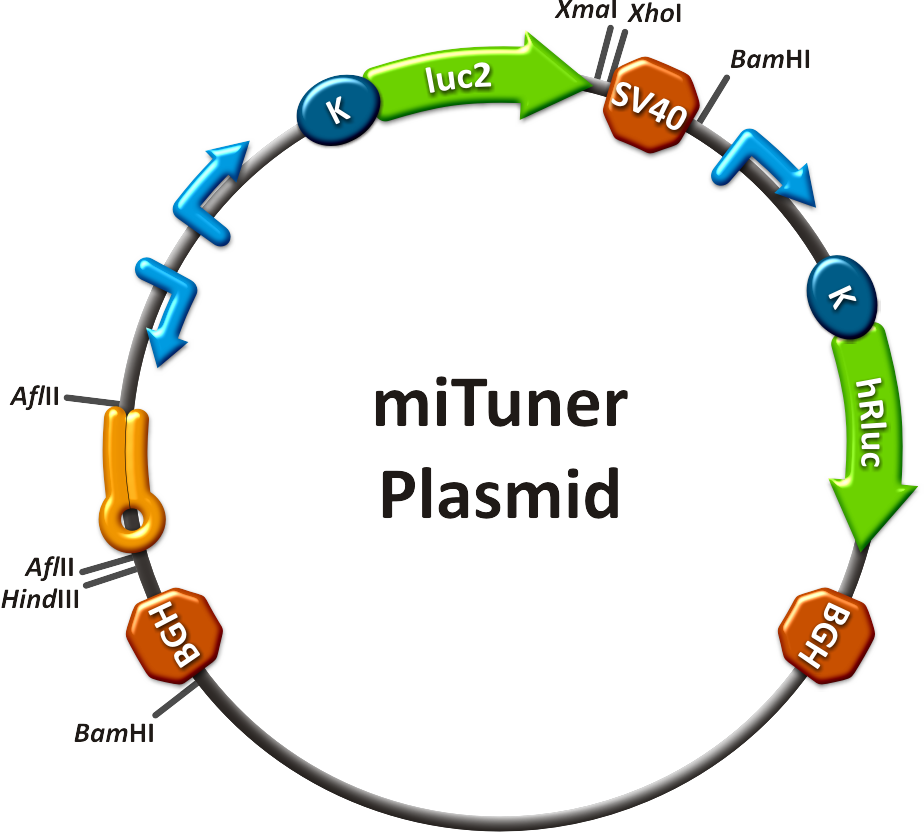
| - | All gene regulatory constructs for tuning, Off- or On-targeting can easily be assembled using '''BBB standard cloning''' from our miRNA Kit [https://2010.igem.org/Team:Heidelberg/Parts parts]. After successful cloning, the constructs can be transfected onto a cell line of choice or transferred into a virus backbone for [https://2010.igem.org/Team:Heidelberg/Project/Mouse_Infection ''in vivo''] experiments. For our ''' | + | All gene regulatory constructs for tuning, Off- or On-targeting can easily be assembled using '''BBB standard cloning''' from our miRNA Kit [https://2010.igem.org/Team:Heidelberg/Parts parts]. After successful cloning, the constructs can be transfected onto a cell line of choice or transferred into a virus backbone for [https://2010.igem.org/Team:Heidelberg/Project/Mouse_Infection ''in vivo''] experiments. For our '''proof of principle''', we used firefly luciferase normalized to ''Renilla'' luciferase on miTuner to characterize knockdown efficiencies of different binding sites and show Off- and On-targeting by mouse infection carried by an AAV virus. |
===miTuner: Expression Fine-Tuning by Synthetic miRNAs=== | ===miTuner: Expression Fine-Tuning by Synthetic miRNAs=== | ||
| Line 56: | Line 56: | ||
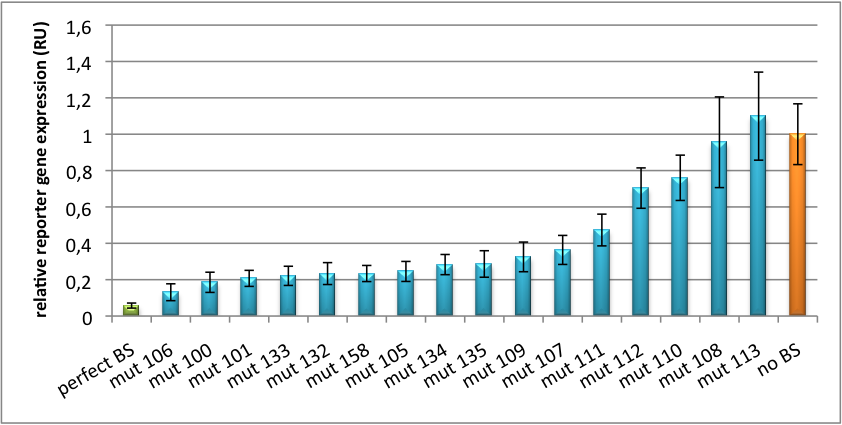
[[Image:PsiCheck.png|thumb|center|600px|'''Figure 3: Tuning of gene expression through different imperfect miR122 binding sites in psiCHECK-2.''' Construct was transfected into HeLa cells together with an plasmid expressing miR122. Control without binding site was used for normalization.]] | [[Image:PsiCheck.png|thumb|center|600px|'''Figure 3: Tuning of gene expression through different imperfect miR122 binding sites in psiCHECK-2.''' Construct was transfected into HeLa cells together with an plasmid expressing miR122. Control without binding site was used for normalization.]] | ||
| + | |||
| + | We further analyized binding sites derived from miR122 in the dual luciferase vector PsiCheck2. Here we tested sixteen mutated binding sites in order to observe minute fine-tuning between one binding site and the next. | ||
===Off-Targeting Using Endogenous miRNA=== | ===Off-Targeting Using Endogenous miRNA=== | ||
Revision as of 17:33, 27 October 2010

|
|
||
 "
"