Team:Heidelberg/Project/miRNA Kit
From 2010.igem.org
Laura Nadine (Talk | contribs) (→miTuner: Expression Fine-Tuning by Synthetic miRNAs) |
Laura Nadine (Talk | contribs) |
||
| Line 57: | Line 57: | ||
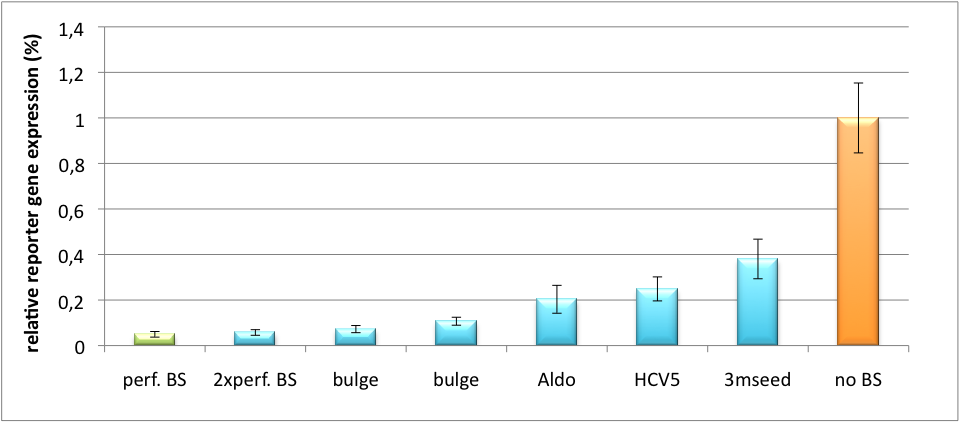
[[Image:PsiCheck.png|thumb|center|600px|'''Figure 3: Tuning of gene expression through different imperfect miR122 binding sites in psiCHECK-2.''' Construct was transfected into HeLa cells together with an plasmid expressing miR122. Control without binding site was used for normalization.]] | [[Image:PsiCheck.png|thumb|center|600px|'''Figure 3: Tuning of gene expression through different imperfect miR122 binding sites in psiCHECK-2.''' Construct was transfected into HeLa cells together with an plasmid expressing miR122. Control without binding site was used for normalization.]] | ||
| - | ===Off-Targeting | + | ===Off-Targeting Using Endogenous miRNA=== |
Another application of our synthetic miRNA Kit profits of tissue specific endogenous miRNAs expression. These can be exploited for either Off- or On-Targeting. | Another application of our synthetic miRNA Kit profits of tissue specific endogenous miRNAs expression. These can be exploited for either Off- or On-Targeting. | ||
| Line 64: | Line 64: | ||
[[Image:HuH Offpng.png|thumb|center|500px|'''Figure 4: Knockdown of reporter gene expression due to endogenous miR122 that interferes with binding sites.''' Construct transfected to HuH cells to off-target those.]] | [[Image:HuH Offpng.png|thumb|center|500px|'''Figure 4: Knockdown of reporter gene expression due to endogenous miR122 that interferes with binding sites.''' Construct transfected to HuH cells to off-target those.]] | ||
| - | ===On-Targeting | + | ===On-Targeting Using Endogenous miRNA=== |
In line with the Off-targeting approach, In the case of On-targeting the presence of a certain miRNA in a cell switches on expression of the GOI. This can be accomplished by using a repressor that is targeted by an endogenously expressed miRNA. We exemplified this scenario by using a Tet Repressor fused with a perfect binding site for miRNA 122, a liver-specific miRNA (REF!). At the same time, the promoter expressing the GOI would be under control of a Tet Operator. Upon presence of the miRNA 122, the Tet Repressor would be knocked down, release the promoter and expression of the GOI could be established. | In line with the Off-targeting approach, In the case of On-targeting the presence of a certain miRNA in a cell switches on expression of the GOI. This can be accomplished by using a repressor that is targeted by an endogenously expressed miRNA. We exemplified this scenario by using a Tet Repressor fused with a perfect binding site for miRNA 122, a liver-specific miRNA (REF!). At the same time, the promoter expressing the GOI would be under control of a Tet Operator. Upon presence of the miRNA 122, the Tet Repressor would be knocked down, release the promoter and expression of the GOI could be established. | ||
| Line 102: | Line 102: | ||
==Methods== | ==Methods== | ||
| - | ===miTuner: Expression | + | ===miTuner: Expression Fine-Tuning by Synthetic miRNAs=== |
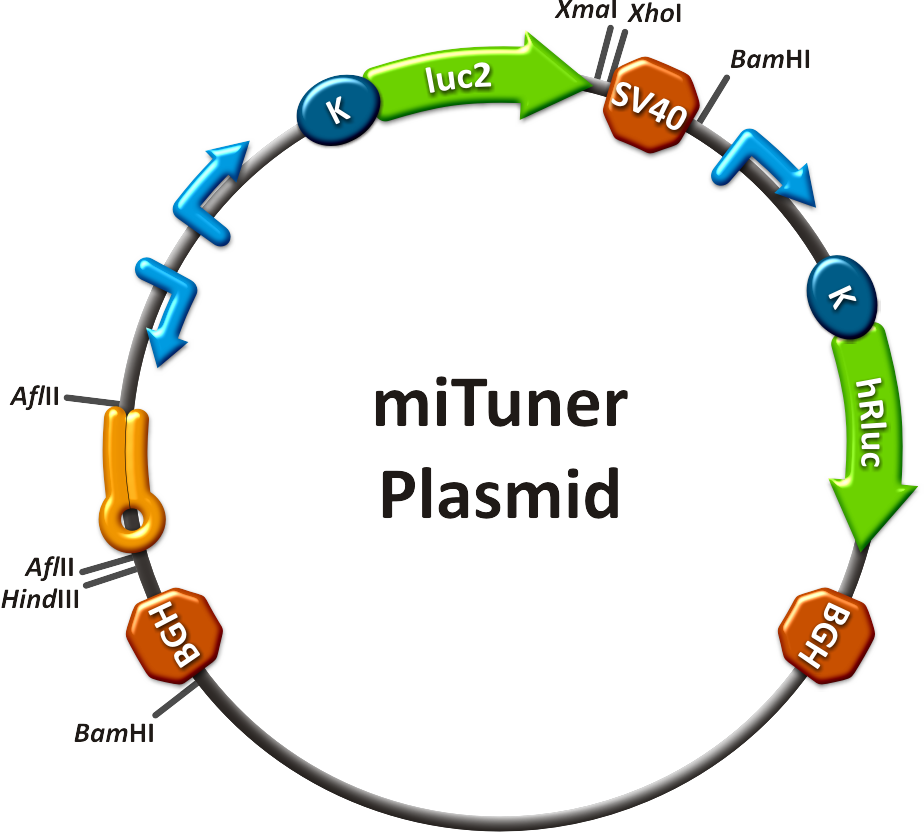
The miTuner was [https://2010.igem.org/3A_Assembly assembled] out of different [https://2010.igem.org/Team:Heidelberg/Parts parts]. Cloning was done following [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Cloning standard protocols].<br> | The miTuner was [https://2010.igem.org/3A_Assembly assembled] out of different [https://2010.igem.org/Team:Heidelberg/Parts parts]. Cloning was done following [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#Cloning standard protocols].<br> | ||
| Line 138: | Line 138: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | === | + | === Working Modes === |
The synthetic miR Kit can be applied in three different ways: | The synthetic miR Kit can be applied in three different ways: | ||
| Line 149: | Line 149: | ||
| - | === | + | === Simple Tuning Procedure === |
* choose an [[Team:Heidelberg/Project/Introduction | interesting microRNA]] | * choose an [[Team:Heidelberg/Project/Introduction | interesting microRNA]] | ||
* [https://2010.igem.org/Team:Heidelberg/Modeling/miBSdesigner create] referring binding sites | * [https://2010.igem.org/Team:Heidelberg/Modeling/miBSdesigner create] referring binding sites | ||
| Line 159: | Line 159: | ||
<br /> | <br /> | ||
| - | === | + | === Advancement === |
* digestion of miR Kit construct with BamHI | * digestion of miR Kit construct with BamHI | ||
* cloning into viral backbone (e. g. [[Team:Heidelberg/Project/Materials/Plasmids | pBS_U6]]) | * cloning into viral backbone (e. g. [[Team:Heidelberg/Project/Materials/Plasmids | pBS_U6]]) | ||
| Line 169: | Line 169: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | === | + | === Tuning Raw Data === |
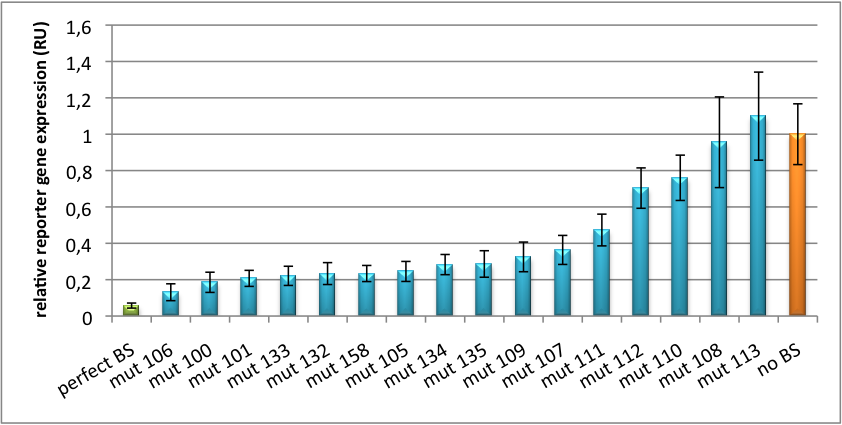
For our <i>in vitro</i> tuning, you can have a look even at our unprocessed data with specific [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#nomenclature nomenclature]: | For our <i>in vitro</i> tuning, you can have a look even at our unprocessed data with specific [https://2010.igem.org/Team:Heidelberg/Notebook/Methods#nomenclature nomenclature]: | ||
* [[Media:Plate1 process H1.xls]], <br/> | * [[Media:Plate1 process H1.xls]], <br/> | ||
Revision as of 17:15, 27 October 2010

|
|
||
 "
"