Team:Tokyo Tech/Project/Artificial Cooperation System
From 2010.igem.org
Contents |
Artificial Cooperation System
Abstract
・We designed Artificial Cooperation System to endow E.coli with humanity.
・We constructed two NEW BioBricks, LuxR repression promoter and chloramphenicol resistance gene connected to the LuxR activation promoter.
・We characterized the existing parts R0061, LuxR repression promoter, and R0062, LuxR activation promoter.
・We characterized the existing part K092400, rbs-luxI-ter.
Introduction
What’s Artificial Cooperation System?
Genetic Circuit
I, genetic circuit overview
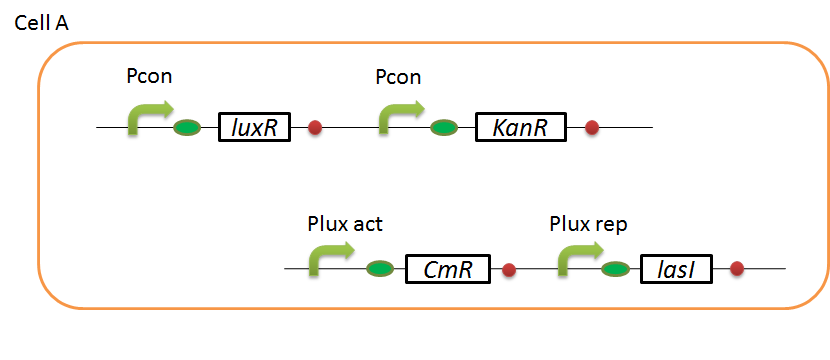
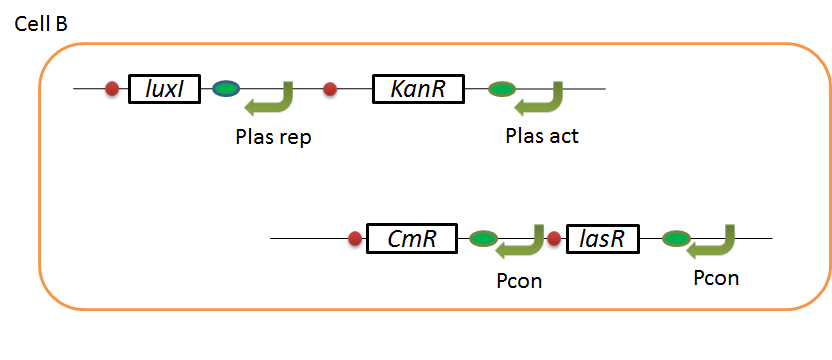
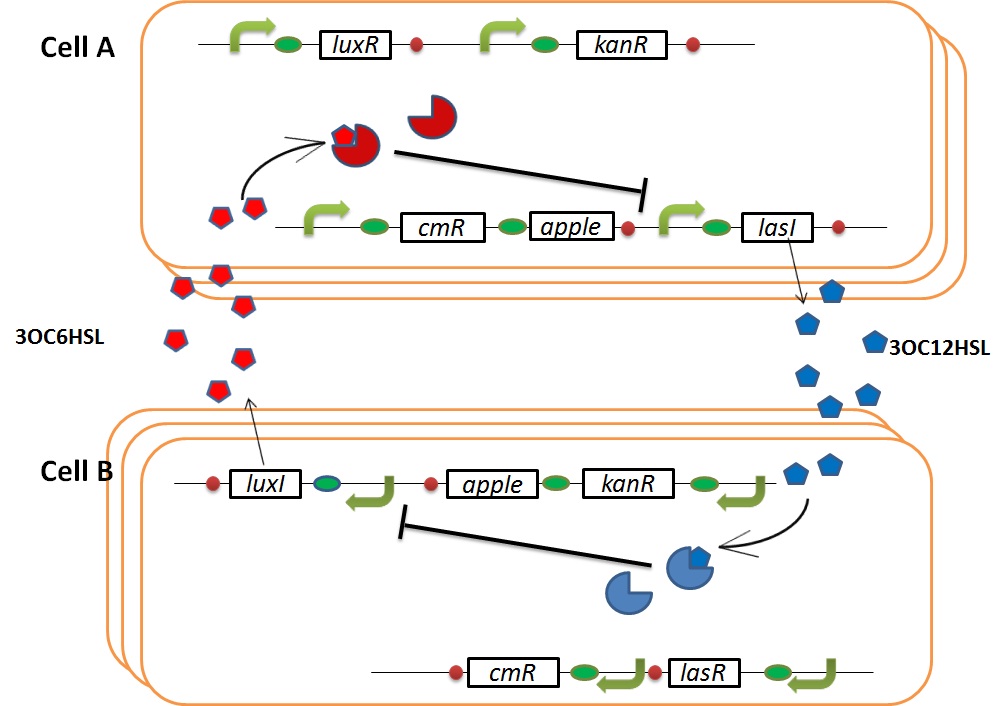
We designed the following circuit. In Artificial Cooperation System, there are two types of E. coli, Cell A and Cell B. Cell A has resistance to kanamycin and Cell B has resistance to chloramphenicol originally.
Pcon: constitutive promoter.
Plux act: promoter activated by LuxR and 3OC6HSL. We call this promoter luxR activation promoter.
Plux rep: promoter repressed by LuxR and 3OC6HSL. We call this promoter luxR repression promoter.
Plas act: promoter activated by LasR and 3OC12HSL. We call this promoter lasR activation promoter.
Plas rep: promoter repressed by LasR and 3OC12HSL. We call this promoter lasR repression promoter.
In cell A, production of LasI is repressed by LuxR/3OC6HSL complex and Cm resistance gene is activated by LuxR/3OC6HSL complex. In cell B, production of LuxI is repressed by LasR/3OC12HSL complex and Kan resistance gene is activated by LasR/3OC12HSL complex. This means AHL of cell A and cell B repress each other indirectly. And AHL of cell A activates resistance gene of cell B, and AHL of cell B activates resistance gene of cell A. Apple gene is composed of crtEBIYZW and MpAAT1. This expresses only when the cell is helped.
Normal
In normal situation, Cell A and Cell B are competitors and recognize each other by using quorum sensing. First, LasI protein in Cell A produces 3OC12HSL. Second, these signal molecules diffuse through cell membranes and bind to LasR protein in Cell B. Third, LasR/3OC12HSL complex represses the production of LuxI protein. Similarly in cell B, LuxI protein produces 3OC6HSL. These signal molecules diffuse through cell membranes and bind to LuxR protein in Cell A. LuxR/OC6HSL complex represses the production of LasI protein. Although Cell A and Cell B have chloramphenicol and kanamycin resistance gene respectively, they don’t express in this situation. That’s because the concentration of 3OC12HSL and 3OC6HSL is too low to activate kanamycin resistance gene and chloramphenicol resistance gene, respectively.
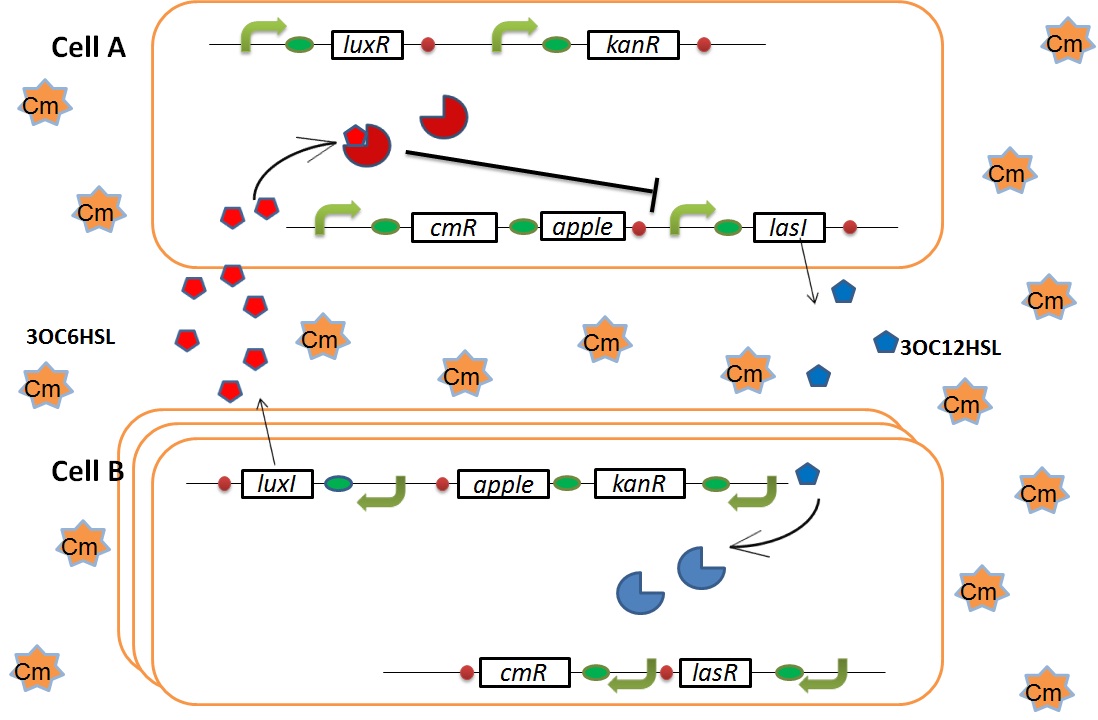
when Cell A is dying
As we mentioned, originally Cell A has resistance to kanamycin and doesn’t have resistance to chloramphenicol. On the other hand, Cell B has resistance to chloramphenicol and doesn’t have resistance to kanamycin. Therefore, number of only Cell A decreases after addition of chloramphenicol. This means 3OC12HSL decreases. And it also results in decreasing of 3OC12HSL and LasR complex in Cell B. By this process, Cell B notices that Cell A is dying and tries to rescue it.
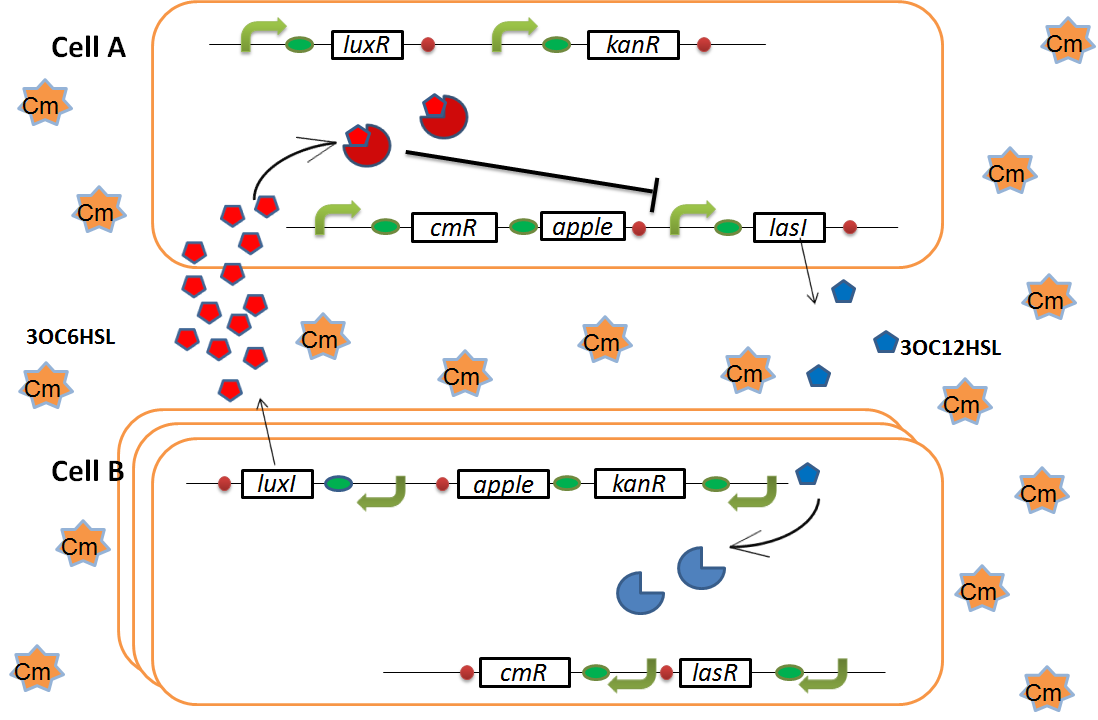
when Cell B tries to rescue Cell A
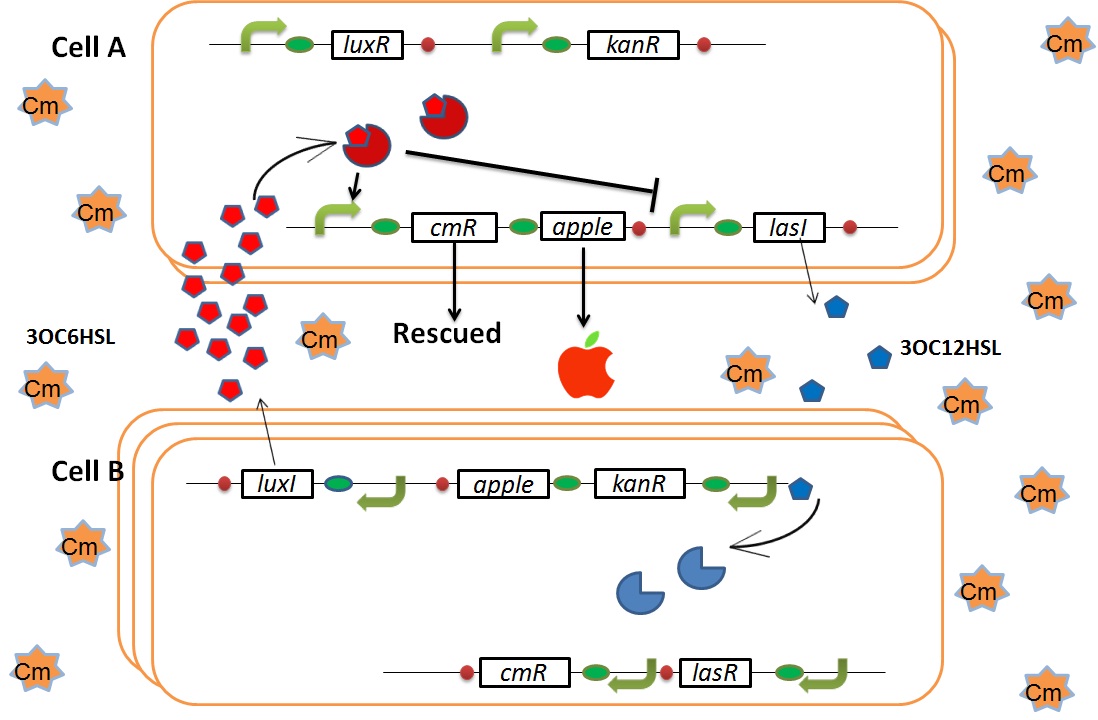
As we mentioned, the expression of luxI is repressed by LasR and 3OC12HSL complex. Therefore, the decrease of 3OC12HSL and LasR complex leads to overexpression of luxI and 3OC6HSL. It results in the activation of chloramphenicol resistance gene and this is followed by increase of the number of Cell A. Consequently, the mission of rescuing Cell A is completed!
II, Promoters repressed by LuxR
In our system, we need promoters repressed by R protein. The fact that LuxR and TraR can function as both activation and repression has already been published in a paper*. We have characterized luxR promoter in order to confirm this feature of LuxR protein.
- Conversion of the Vibrio fischeri Transcriptional Activator, LuxR, to a Repressor
III, Tuning of strength and threshold of promoters
To make our Artificial Cooperative System work, strength and threshold of promoters regulated by AHL are so important. Therefore we have to tune threshold and strength of promoters. We tried tuning promoters by changing sequence of consensus -35/-10 and lux box. See more
Result
私たちは助け合うシステムの実現のためexisting partsnであるLuxR repression promoter R0061とLuxR activation promoter R0062のcharacterizationのための実験。あらたに作ったLuxR repression promoter K395008 の作成およびcharacterization
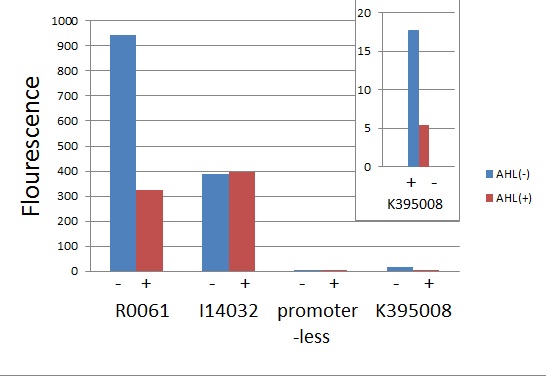
LuxR repression promoter and activation promoter
We characterized LuxR repression promoters and activation promoters, because in the Artificial Cooperation System, both LuxR repression promoter and activation promoter plays important roles. The following figures are the result of experiments. Fig〇〇 shows that R0061 and K395008 work as we expected. R0061 is existing BioBrick LuxR repression promoter and K395008 is LuxR repression promoter we designed. Fig〇〇 shows that R0062,which is existing LuxR activation promoter, works as we expected. Therefore we characterized existing BioBrick parts, R0061 and R001 and succeeded in making a new LuxR repression promoter, K395008. See more
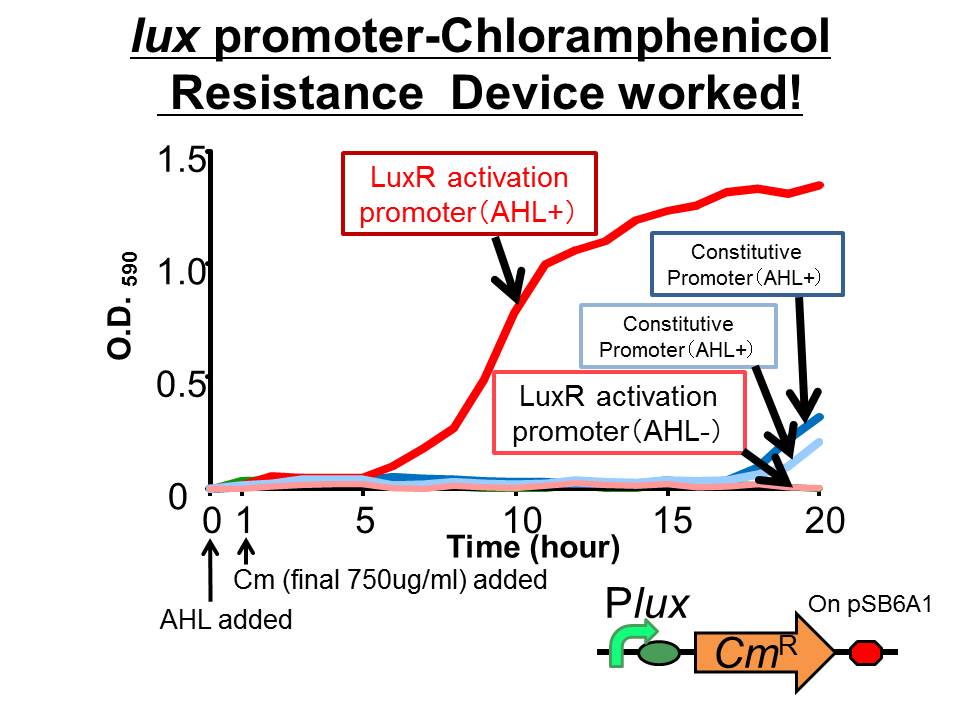
plux-CmR
We succeeded in constructing and characterizing a NEW Biobrick device of chloramphenicol resistance (CmR) gene ([http://partsregistry.org/Part:BBa_K395162 BBa_K395162]) which is activated by lux promoter. Fig〇〇 showsthat the device is activated by LuxR/3OC6HSL complex and that the cell introduced the part was able to survive even in high chloramphenicol concentration (750 ug/ml) in the presence of 3OC6HSL. See more
luxI
Mathematical modeling
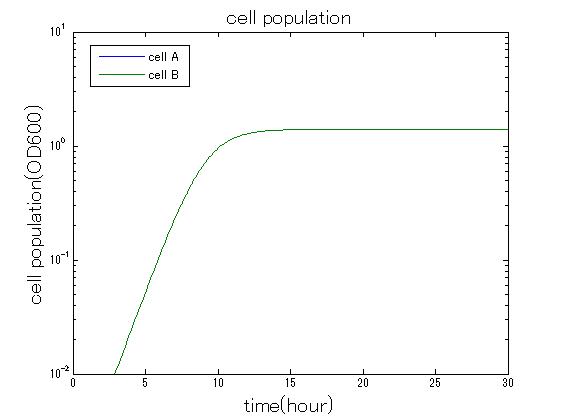
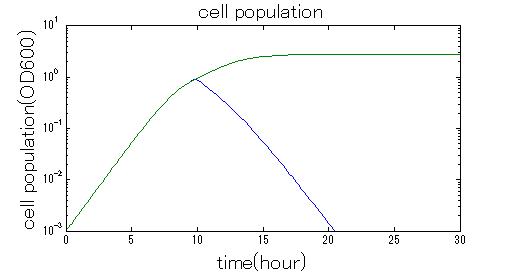
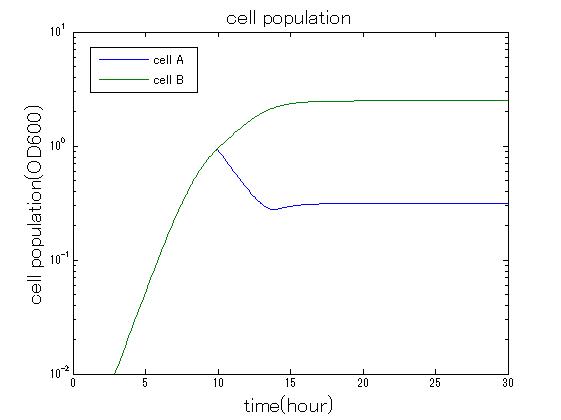
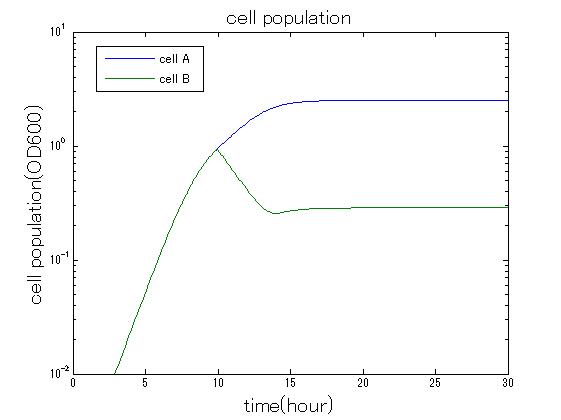
In order to confirm that this system is feasible, we simulated this system. The following figures 〇〇 show the simulation result. We held a series of simulations under several experimental conditions. Fig◯◯〜Fig◯◯ are the results. Fig1 is the case without antibiotic. In this case, cell A and cell B show no difference of growth. Fig2 is the condition with chloramphenicol while ACS is not working. Graph shows that cellA decrease by chloramphenicol. Fig.3 is the condition with chloramphenicol while ACS is working. The graph shows turnover of CellA, which means it was rescued by ACS. Fig4 is the condition with Kanamycin while ACS is working. This time, number of CellB decrease at first, though recover at the end. From the simulation results above, we can conclude that our system can work under a certain situation.
 "
"