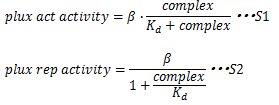Team:Tokyo Tech/Project/Artificial Cooperation System
From 2010.igem.org
(→Genetic circuit) |
(→Works) |
||
| Line 82: | Line 82: | ||
In these equations,βrepresents Plux max activity, complex represents the concentration of LuxR/3OC6HSL complex and Kd represents the affinity of LuxR and lux box. S1 and S2 represents that Kd doesn’t determine the Plux max activity, but determines the complex concentration producing half level from this promoter. So, changing sequence of lux box doesn’t lead changing the Plux max activity. This leads changing threshold of Plux. | In these equations,βrepresents Plux max activity, complex represents the concentration of LuxR/3OC6HSL complex and Kd represents the affinity of LuxR and lux box. S1 and S2 represents that Kd doesn’t determine the Plux max activity, but determines the complex concentration producing half level from this promoter. So, changing sequence of lux box doesn’t lead changing the Plux max activity. This leads changing threshold of Plux. | ||
β represents Plux max activity, thus β represents the strength of -35 and -10 sequence. Therefore changing sequence of -35 and -10 leads changing Plux max activity. | β represents Plux max activity, thus β represents the strength of -35 and -10 sequence. Therefore changing sequence of -35 and -10 leads changing Plux max activity. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Revision as of 09:53, 26 October 2010
Contents |
Artificial Cooperation System
Highlight
・We designed Artificial Cooperation System to endow E.coli with humanity.
・We constructed two NEW BioBricks, LuxR repression promoter and chloramphenicol resistance gene connected to the LuxR activation promoter.
・We characterized the existing parts R0061, LuxR repression promoter, and R0062, LuxR activation promoter.
・We characterized the existing part K092400, rbs-luxI-ter.
Requirement
In order to make our system, we need introduce cell-to-cell communication mechanism. Many cell-to-cell communication mechanisms in bacteria are known, though the mechanisms of many systems are not well-understood yet. Thus we used “quorum sensing” whose mechanism is well studied. And quorum sensing is popular in synthetic biology and iGEM.
quorum sensing
Quorum Sensing (QS) refers to cell-to-cell communication systems that are used by many microorganisms to sense their local cell densities. This sensing mechanism is based on the production, secretion and detection of small signaling molecules, whose concentration correlates to the abundance of secreting microorganisms in the vicinity. When the signal concentration reaches a threshold, they know the ‘quorum’ is formed, and the communicating microorganisms undergo a coordinated change in their gene-expression profiles. Secretion of virulence factors, initiation of biofilm formation, sporulation, competence, mating, root nodulation, bioluminescence, and production of secondary metabolites are the examples mentioned above.
There are two regulatory genes involved in QS, I and R genes. The I gene produces I protein and directs the synthesis of an N-acyl homoserine lactone (AHL). AHL is bacteria-specific and gene-specific signal molecule. On the other hand, R protein, which is expressed by R gene, binds to the synthesized AHL signal molecule. The AHL and R protein complex acts as a transcription factor.
In synthetic biology field or iGEM, lux, las, and rhl have been used frequently.lux, lasI and rhlI gene synthesize N-(3-oxohexanoyl) homoserine lactone(3OC6HSL), 3-oxododecanoyl- homoserine lactone (3OC12HSL) and butanoyl- homoserine lactone (C4HSL) ,respectively. LuxR, LasR and RhlR receptor have been synthesized and can bind to 3OC6HSL, 3OC12HSL and C4HSL, respectively.
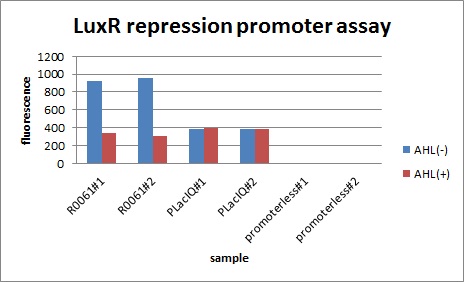
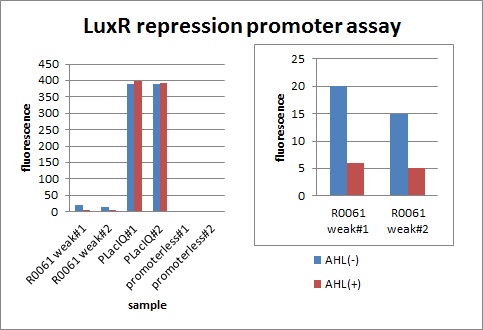
In QS, not only activation promoters but also repression promoters have been used. The fact that LuxR and TraR promoter can function as both activation and repression has already been published in a paper*. We have characterized luxR promoter in order to confirm this feature of LuxR protein.
- Conversion of the Vibrio fischeri Transcriptional Activator, LuxR, to a Repressor
Genetic circuit
I, genetic circuit overview
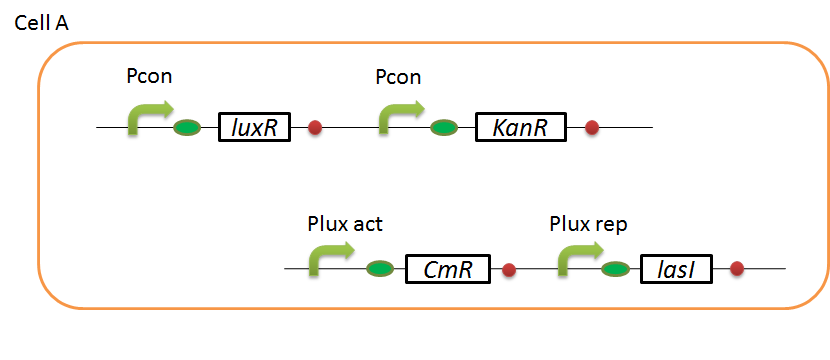
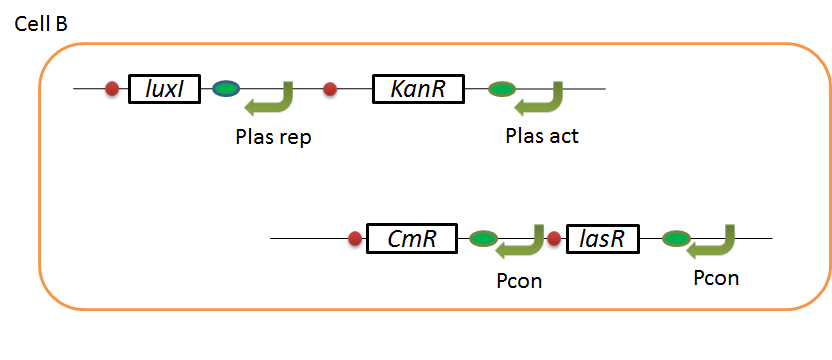
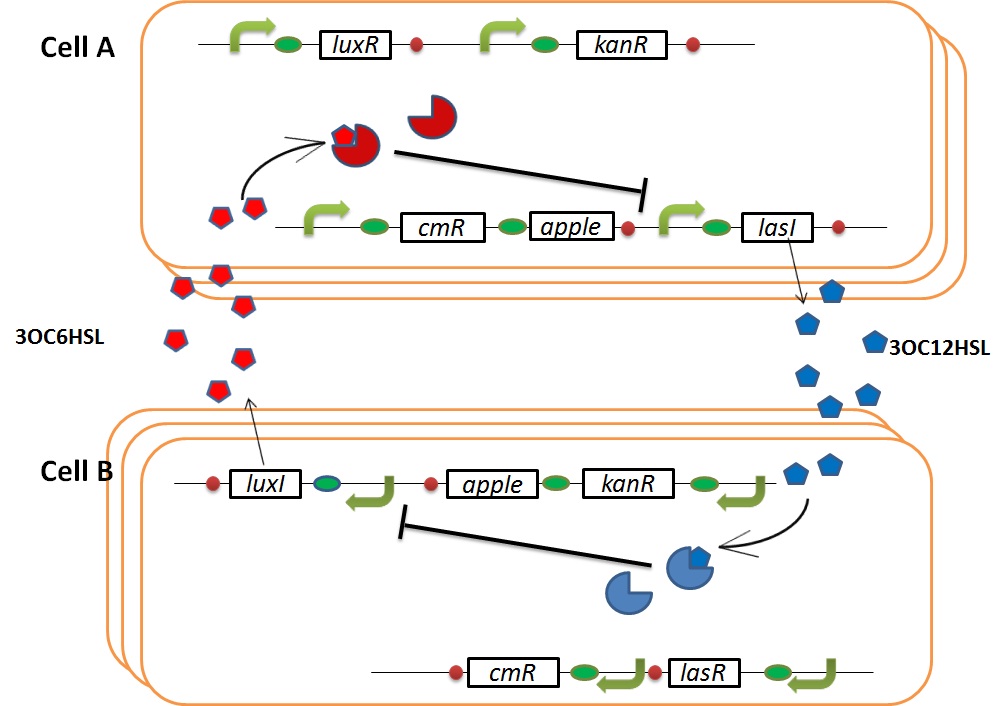
We designed the following circuit. In Artificial Cooperation System, there are two types of E. coli, Cell A and Cell B. Cell A has resistance to kanamycin and Cell B has resistance to chloramphenicol originally.
Pcon: constitutive promoter.
Plux act: promoter activated by LuxR and 3OC6HSL. We call this promoter luxR activation promoter.
Plux rep: promoter repressed by LuxR and 3OC6HSL. We call this promoter luxR repression promoter.
Plas act: promoter activated by LasR and 3OC12HSL. We call this promoter lasR activation promoter.
Plas rep: promoter repressed by LasR and 3OC12HSL. We call this promoter lasR repression promoter.
In cell A, production of LasI is repressed by LuxR/3OC6HSL complex and Cm resistance gene is activated by LuxR/3OC6HSL complex. In cell B, production of LuxI is repressed by LasR/3OC12HSL complex and Kan resistance gene is activated by LasR/3OC12HSL complex. This means AHL of cell A and cell B repress each other indirectly. And AHL of cell A activates resistance gene of cell B, and AHL of cell B activates resistance gene of cell A. Apple gene is composed of crtEBIYZW and MpAAT1. This expresses only when the cell is helped.
normal
In normal situation, Cell A and Cell B are competitors and recognize each other by using quorum sensing. First, LasI protein in Cell A produces 3OC12HSL. Second, these signal molecules diffuse through cell membranes and bind to LasR protein in Cell B. Third, LasR/3OC12HSL complex represses the production of LuxI protein. Similarly in cell B, LuxI protein produces 3OC6HSL. These signal molecules diffuse through cell membranes and bind to LuxR protein in Cell A. LuxR/OC6HSL complex represses the production of LasI protein. Although Cell A and Cell B have chloramphenicol and kanamycin resistance gene respectively, they don’t express in this situation. That’s because the concentration of 3OC12HSL and 3OC6HSL is too low to activate kanamycin resistance gene and chloramphenicol resistance gene, respectively.
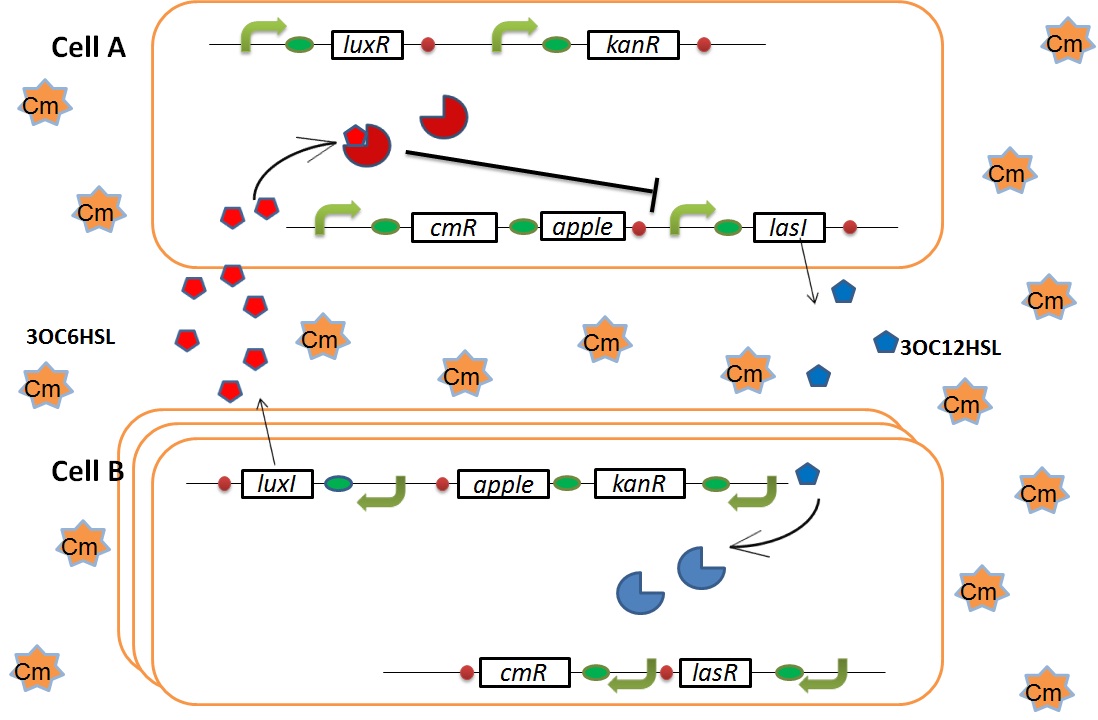
when Cell A is dying
As we mentioned, originally Cell A has resistance to kanamycin and doesn’t have resistance to chloramphenicol. On the other hand, Cell B has resistance to chloramphenicol and doesn’t have resistance to kanamycin. Therefore, number of only Cell A decreases after addition of chloramphenicol. This means 3OC12HSL decreases. And it also results in decreasing of 3OC12HSL and LasR complex in Cell B. By this process, Cell B notices that Cell A is dying and tries to rescue it.
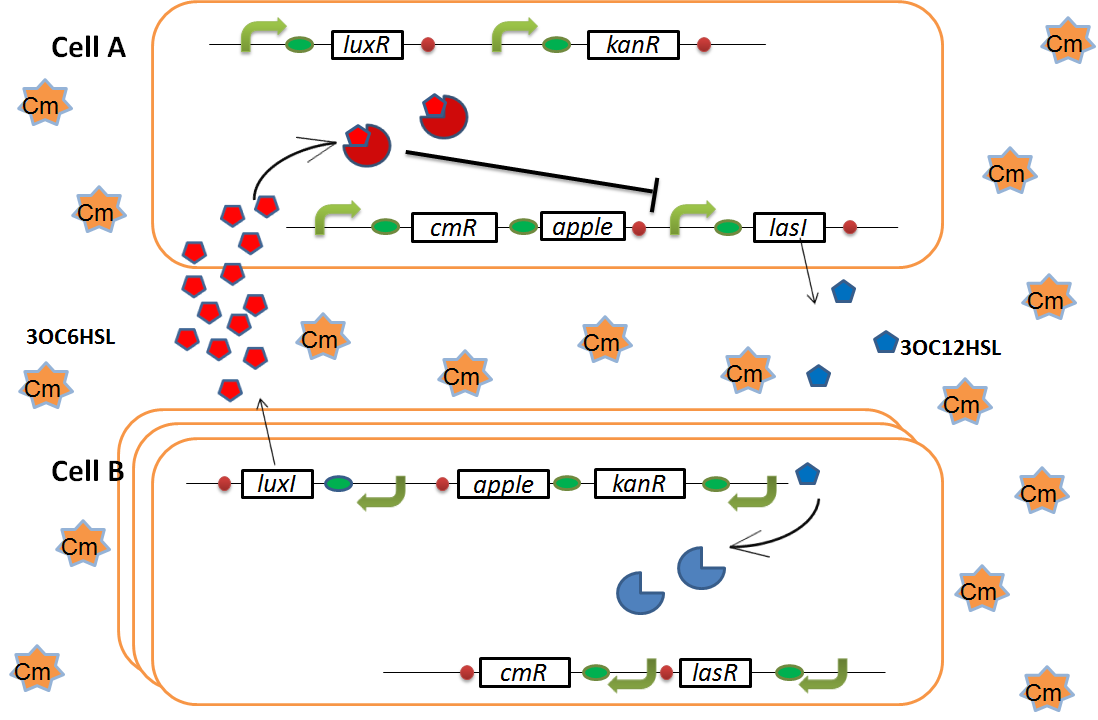
when Cell B tries to rescue Cell A
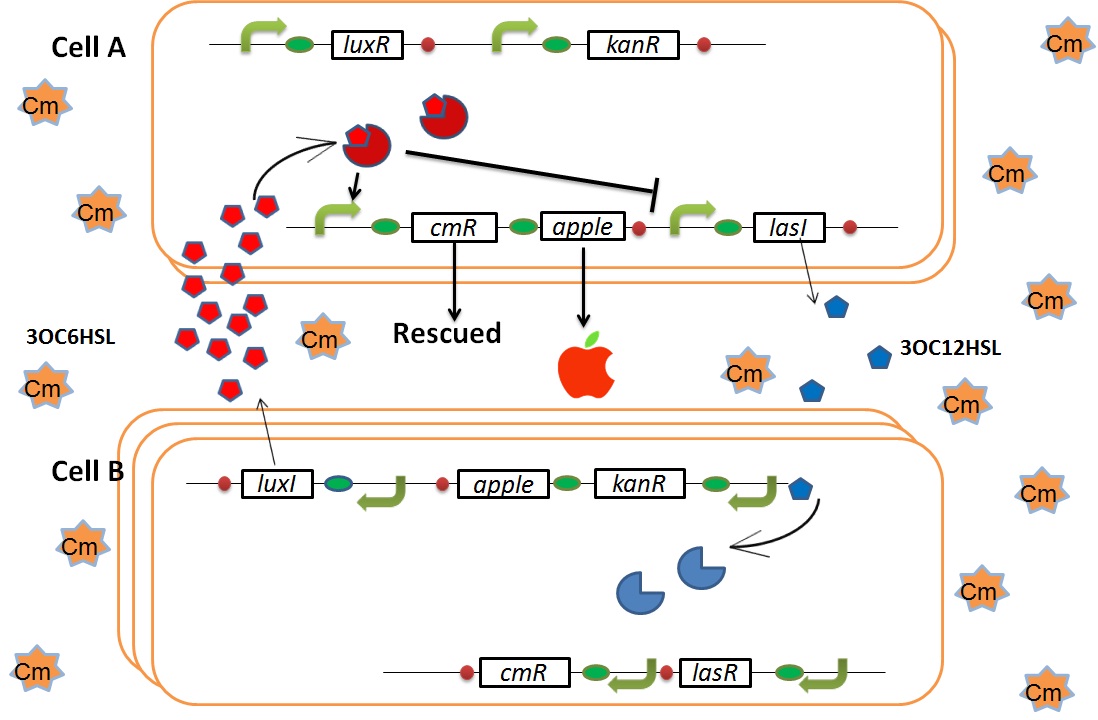
As we mentioned, the expression of luxI is repressed by LasR and 3OC12HSL complex. Therefore, the decrease of 3OC12HSL and LasR complex leads to overexpression of luxI and 3OC6HSL. It results in the activation of chloramphenicol resistance gene and this is followed by increase of the number of Cell A. Consequently, the mission of rescuing Cell A is completed!
II, importance of strength and threshold of promoters
To make our Artificial Cooperative System work, strength and threshold of promoters regulated by AHL are so important. For example, if the threshold of the resistance gene promoter is very low, resistance gene is activated by low AHL level. This means we can’t make a dangerous situation. And if the threshold of promoter of I gene is so high, the production level of I protein stays at highest level all the time. This means two types of cell help each other all the time. Even if the threshold of promoters is suitable, our system doesn’t work unless the strength of promoter is suitable. For example, when the strength of resistance gene promoter is low, cells can’t express adequate resistance gene and die. And if the strength of I protein promoter is low, cell can’t produce adequate amount of AHL to help the other cell. Therefore, we have to design promoters whose threshold and strength are suitable for the Artificial Cooperative System.
III, tuning the threshold and strength of promoter
To initiate transcription from Plux, LuxR needs to bind the regulatory region upstream of -35, lux box. In the absence of AHL, LuxR can’t bind lux box and can’t initiate transcription from Plux. However in the presence of AHL, AHL binds LuxR and then, LuxR/AHL complex can bind lux box and initiate transcription from Plux. Thus, how efficiently transcription initiate from Plux is determined by not only the concentration of LuxR/AHL, but the concentration of LuxR/AHL binding lux box. This concentration is determined by not only AHL, LuxR and LuxR/AHL concentration but also the binding affinity of LuxR/AHL complex and lux box. If the affinity is low, high concentration of LuxR/AHL complex is needed to initiate transcription efficiently. But if the affinity is high, high concentration of LuxR/AHL complex is not needed to initiate transcription efficiently. Therefore we can tune the AHL threshold of Plux by changing the sequence of lux box. These are represented by the following equations. And we can tune the strength of promoters by changing the sequence of -35 and -10 consensus sequence. Therefore we can obtain our ideal promoters by changing sequence.
In these equations,βrepresents Plux max activity, complex represents the concentration of LuxR/3OC6HSL complex and Kd represents the affinity of LuxR and lux box. S1 and S2 represents that Kd doesn’t determine the Plux max activity, but determines the complex concentration producing half level from this promoter. So, changing sequence of lux box doesn’t lead changing the Plux max activity. This leads changing threshold of Plux.
 "
"