Team:Newcastle/12 August 2010
From 2010.igem.org
(→Conclusion) |
(→Conclusion) |
||
| (3 intermediate revisions not shown) | |||
| Line 16: | Line 16: | ||
!'''Tube''' | !'''Tube''' | ||
!'''Part to be amplified''' | !'''Part to be amplified''' | ||
| - | !'''DNA | + | !'''Template DNA''' |
!'''Forward primer''' | !'''Forward primer''' | ||
!'''Reverse Primer''' | !'''Reverse Primer''' | ||
!'''Melting Temperature (Tm in °C) ''' | !'''Melting Temperature (Tm in °C) ''' | ||
!'''Size of the fragment (in bp)''' | !'''Size of the fragment (in bp)''' | ||
| - | !'''Extension time | + | !'''Extension time (in seconds)''' |
|- | |- | ||
|1 | |1 | ||
| Line 88: | Line 88: | ||
==Discussion== | ==Discussion== | ||
| - | We found that the PCR tube containing the ''spaIFEG'' gene (part 3) did not have any band, even though another gel electrophoresis was performed (at 63°C) again. We decided to perform five different PCR reactions at different temperatures: 46°C, 51°C, 56°C, 61°C and 66°C - this is to check whether the missing band was due to the incorrect Tm. We also performed a sixth PCR tube as a control, which contained the ''ara'' forward and reverse primers (we previously had extracted from ''Bacillus subtilis'' ATCC 6633 on [[Team:Newcastle/7_July_2010#Chromosomal_prep| 7th July 2010]]) - the melting temperature for this was 59°C, with the extention time of fifteen seconds (specific for ''ara'' primers). ''ara'' primers were put into the tube because they were the primers that confirmed that the chromosomal DNA extraction from ''Bacillus subtilis'' ATCC 6633 had worked. | + | We found that the PCR tube containing the ''spaIFEG'' gene (part 3) did not have any band, even though another gel electrophoresis was performed (at 63°C) again. We decided to perform five different PCR reactions at different temperatures: 46°C, 51°C, 56°C, 61°C and 66°C - this is to check whether the missing band was due to the incorrect Tm. We also performed a sixth PCR tube as a control, which contained the ''ara'' forward and reverse primers (we previously had extracted from ''Bacillus subtilis'' ATCC 6633 on [[Team:Newcastle/7_July_2010#Chromosomal_prep| 7th July 2010]]) - the melting temperature for this was 59°C, with the extention time of fifteen seconds (specific for ''ara'' primers). The ''ara'' primers were put into the tube because they were the primers that confirmed that the chromosomal DNA extraction from ''Bacillus subtilis'' ATCC 6633 had worked. |
We have cut the Plasmid Vector (part 1), Promoter & RBS (part 2) and Double terminator (part 4) gel bands following the [[Team:Newcastle/Gel_extraction| gel extraction]] protocol. We will wait until the ''spaIFEG'' gene (part 3) has been cut from the gel. This will be carried out [[Team:Newcastle/12 August 2010#Subtillin_immunity_BioBrick| tommorrow]]. After all four parts have been cut from gels, the[[Team:Newcastle/Gel_extraction| gel extraction]] protocol will be completed and the concentration of the DNA will be tested using the [[TeamNewcastleNanoDrop_Spectrophotometer| NanoDrop Spectrophotometer]]. Once all four parts have been successfully extracted, a single PCR tube will be prepared for the final step for the Gibson cloning. | We have cut the Plasmid Vector (part 1), Promoter & RBS (part 2) and Double terminator (part 4) gel bands following the [[Team:Newcastle/Gel_extraction| gel extraction]] protocol. We will wait until the ''spaIFEG'' gene (part 3) has been cut from the gel. This will be carried out [[Team:Newcastle/12 August 2010#Subtillin_immunity_BioBrick| tommorrow]]. After all four parts have been cut from gels, the[[Team:Newcastle/Gel_extraction| gel extraction]] protocol will be completed and the concentration of the DNA will be tested using the [[TeamNewcastleNanoDrop_Spectrophotometer| NanoDrop Spectrophotometer]]. Once all four parts have been successfully extracted, a single PCR tube will be prepared for the final step for the Gibson cloning. | ||
==Conclusion== | ==Conclusion== | ||
| - | We have successfully extracted part 3 (''spaIFEG'' Gene Cluster) | + | We have successfully extracted part 3 (''spaIFEG'' Gene Cluster). |
| - | + | ||
Latest revision as of 23:28, 27 October 2010

| |||||||||||||
| |||||||||||||
Contents |
PCR amplification of subtilin immunity BioBrick fragments
Aims
The aim for this experiment is to PCR amplify the spaIFEG gene cluster using a range of melting temperature.
Materials and protocol
Please refer to the gel electrophoresis and the PCR protocols.
| Tube | Part to be amplified | Template DNA | Forward primer | Reverse Primer | Melting Temperature (Tm in °C) | Size of the fragment (in bp) | Extension time (in seconds) |
|---|---|---|---|---|---|---|---|
| 1 | spaIFEG Gene Cluster | B. subtilis ATCC 6633 | P1S1 forward | P2S1 reverse | 46 | 2753 + | 110 |
| 2 | spaIFEG Gene Cluster | B. subtilis ATCC 6633 | P1S1 forward | P2S1 reverse | 51 | 2753 + | 110 |
| 3 | spaIFEG Gene Cluster | B. subtilis ATCC 6633 | P1S1 forward | P2S1 reverse | 56 | 2753 + | 110 |
| 4 | spaIFEG Gene Cluster | B. subtilis ATCC 6633 | P1S1 forward | P2S1 reverse | 61 | 2753 + | 110 |
| 5 | spaIFEG Gene Cluster | B. subtilis ATCC 6633 | P1S1 forward | P2S1 reverse | 66 | 2753 + | 110 |
Table 1: Shows the PCR condition
Result
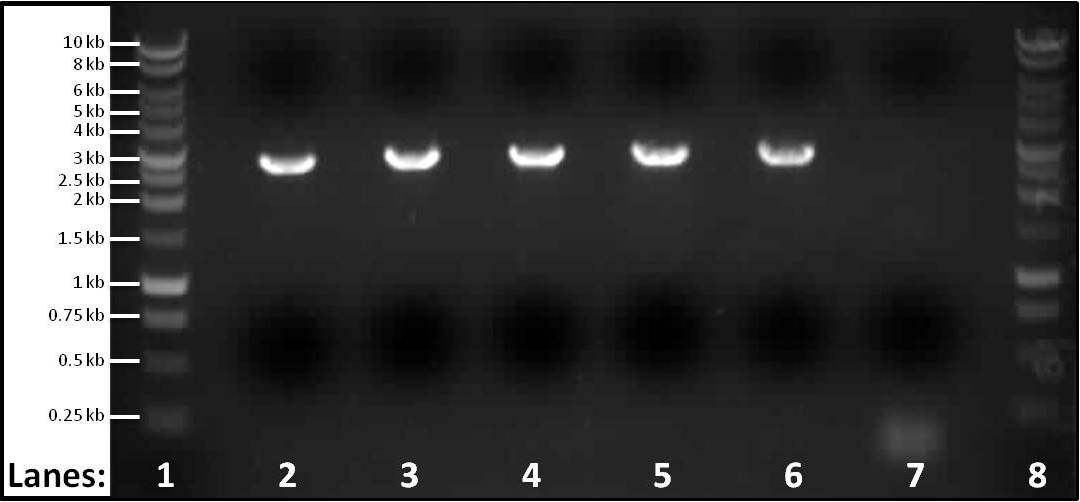
Figure 1: Gel electrophoresis of the PCR products of part 3 spaIFEG gene cluster required for the subtilin immunity BioBrick. The PCR reactions were performed for 5 different melting temperatures. The primers used for lanes 2-6 were the primer 1-S1for(B) and primer 2-S1rev(B). For the control the ara forward and reverse primers were used.
- Lane 1: 1 kb DNA ladder
- Lane 2: spaIFEG Gene Cluster - Tm: 46°C, extension time: 110 seconds
- Lane 3: spaIFEG Gene Cluster - Tm: 51°C, extension time: 110 seconds
- Lane 4: spaIFEG Gene Cluster - Tm: 56°C, extension time: 110 seconds
- Lane 5: spaIFEG Gene Cluster - Tm: 61°C, extension time: 110 seconds
- Lane 6: spaIFEG Gene Cluster - Tm: 66°C, extension time: 110 seconds
- Lane 7: Control - Tm of 59°C, extension time 15 seconds
- Lane 8: 1 kb DNA ladder
Discussion
We found that the PCR tube containing the spaIFEG gene (part 3) did not have any band, even though another gel electrophoresis was performed (at 63°C) again. We decided to perform five different PCR reactions at different temperatures: 46°C, 51°C, 56°C, 61°C and 66°C - this is to check whether the missing band was due to the incorrect Tm. We also performed a sixth PCR tube as a control, which contained the ara forward and reverse primers (we previously had extracted from Bacillus subtilis ATCC 6633 on 7th July 2010) - the melting temperature for this was 59°C, with the extention time of fifteen seconds (specific for ara primers). The ara primers were put into the tube because they were the primers that confirmed that the chromosomal DNA extraction from Bacillus subtilis ATCC 6633 had worked.
We have cut the Plasmid Vector (part 1), Promoter & RBS (part 2) and Double terminator (part 4) gel bands following the gel extraction protocol. We will wait until the spaIFEG gene (part 3) has been cut from the gel. This will be carried out tommorrow. After all four parts have been cut from gels, the gel extraction protocol will be completed and the concentration of the DNA will be tested using the NanoDrop Spectrophotometer. Once all four parts have been successfully extracted, a single PCR tube will be prepared for the final step for the Gibson cloning.
Conclusion
We have successfully extracted part 3 (spaIFEG Gene Cluster).
Go back to our main Lab book page
 
|
 "
"