Team:Calgary/23 June 2010
From 2010.igem.org
(Difference between revisions)
| Line 8: | Line 8: | ||
Today, I did a restriction digest in order to construct the ''cpxP'' PCR product into pSB1A3 plasmid. This was done with ''XbaI'' and ''SpeI'' as those were the only sites on the PCR product. This will result in unintentional scars, but we will hopefully deal with this later on. Along with this, I also constructed an R0040 promoter in front of both E0430 and E0420 (switched onto pSB1K2). | Today, I did a restriction digest in order to construct the ''cpxP'' PCR product into pSB1A3 plasmid. This was done with ''XbaI'' and ''SpeI'' as those were the only sites on the PCR product. This will result in unintentional scars, but we will hopefully deal with this later on. Along with this, I also constructed an R0040 promoter in front of both E0430 and E0420 (switched onto pSB1K2). | ||
| + | |||
| + | '''Dev''' | ||
| + | |||
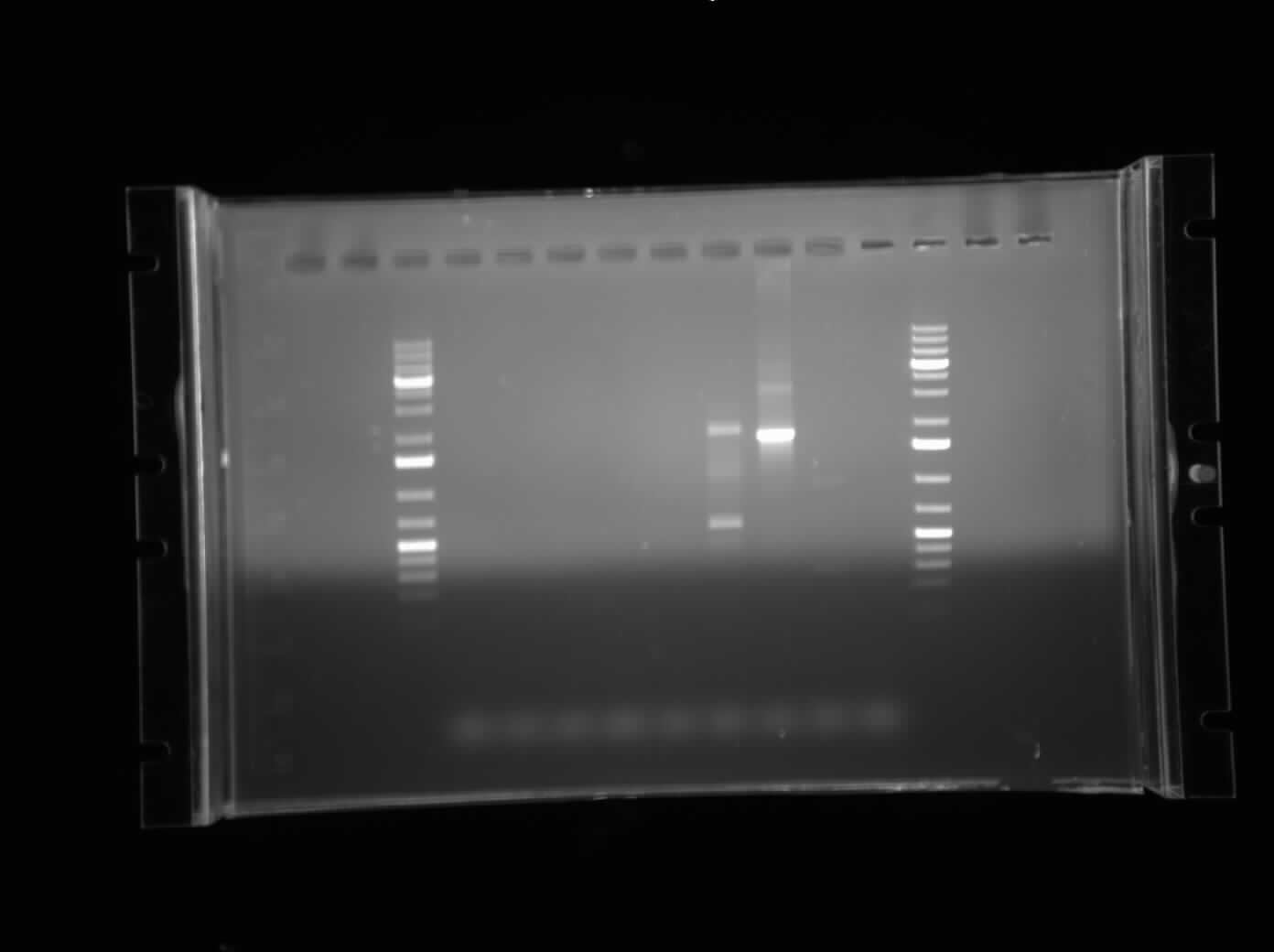
| + | 10 colonies of growth were found from the transformation of the E0040 + B0015 construct from yesterday. A colony PCR was conducted with all 10 colonies. The PCR products were mixed with loading dye and water and ran on a 1% agarose gel. | ||
}} | }} | ||
Revision as of 20:44, 7 July 2010

Wednesday June 23, 2010
Patrick
Today, I did a restriction digest in order to construct the cpxP PCR product into pSB1A3 plasmid. This was done with XbaI and SpeI as those were the only sites on the PCR product. This will result in unintentional scars, but we will hopefully deal with this later on. Along with this, I also constructed an R0040 promoter in front of both E0430 and E0420 (switched onto pSB1K2).
Dev
10 colonies of growth were found from the transformation of the E0040 + B0015 construct from yesterday. A colony PCR was conducted with all 10 colonies. The PCR products were mixed with loading dye and water and ran on a 1% agarose gel.
No notebook page exists for this date. Sorry! "
"