Team:Calgary/10 August 2010
From 2010.igem.org

Tuesday August 10, 2010
Jeremy
Today I ligated the products of pRFP that I had done a restriction digest and antarctic phosphatase on. And also transformed and plated them. I also did a gradient PCR with the remaining amount of pRFP and am doing a colony PCR of the plates where pRFP was gained from as well as the growth from one of the plates from a previous plasmid switch. I ran all the PCR products on a gel and used the information from the colony PCR to do overnights as well as PCR purify the gradient PCR.
Emily
Today I miniprepped all of my malE and malE31 cultures from yesterday. I then set them up to be digested with the remaining enzymes that they were not cut with. If these genes are indeed biobricked, then we would expect to see, when run on an agarose gel, 1 band at ~1200 base pairs for the insert, as well as one band at ~ 3kb for the psB1AC3 and psB1AK3 vectors. I ran a1% gel of my two gradient PCR's from last night, however I got no amplification for either of them. For some reason, our primers are not able to amplify malE31SSdel. Raida proceeded by setting up a PCR reaction with PFU instead of Taq in order to try to get our stuff cloned into TOPO TA vectors. I also ran a gel of the colony PCR that Raida set up last night in order to verify our hypothetical I0500-I13504 and I0500-I13507 constructs. This was using the BBK-CP primers. I ran this on a 1% agarose gel and there were bands in some lanes at the expected size of ~2.2 KB for the IO500-I13504 construct. The bands for the lane of I500-I13507 showed a band around 1kb, indicating that we were not able to get the two pieces together. I proceeded by making overnight cultures of five of the colonies of I0500-I13504 that seemed to have both pieces. I also made overnight cultures of some new colonies of malE31delSS as well as of malE31 X/S C2 in order to make glycerol stocks tomorrow. I also helped Chris make TOP10 Competent Cells.
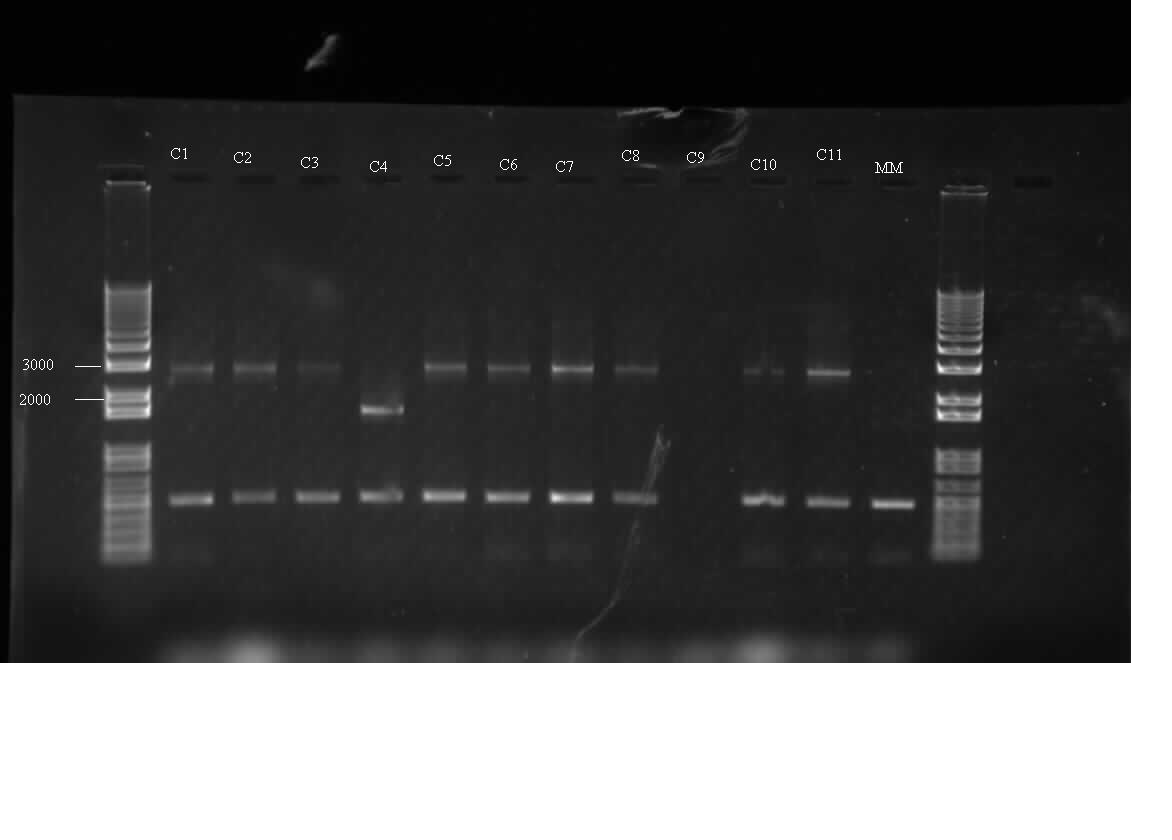
Himika
Today I got growth from the construction that I did last night of I0500-B0034-MalE31. I did a colony PCR of 11 colonies from 4 plates. I used Bbk-CP-F and Bbk-CP-R primers for the amplification. The expected bands were 2600 bp using these primers. The bands that I got on the gel were >2000 and <3000 bp. Fortunately, these were the bands that I got when I ran a gel of the PC product. There were non-specific bands at 500 bp. These 500bp bands were found in the negative control as well. Therefore, these bands are likely due to contamination of the PCR master mix made. I also set up overnight cultures of 5 colonies out of the 11 that I PCR-ed. C1,C2,C5,C7,C11. I also worked on modelling component of the project. I was able to produce a graph using MATLAB. The graph did predict some of the relationships that I was trying to model. The model and relationships do need to be refined further.
Chris
Today, I transformed the parts I13504 and I13507 from the Registry dry DNA. They were transformed into Top10 Competent cells as well as Henry's electroporated cells. I also set up a colony PCR of 16 colonies of CpxP in Ampicillin-kanamycin and cut with either XbaI and SpeI. Overnight cultures were also made of the colonies that were run in the colony PCR in the interest of time. Finally, I finished making more Top10 Competent cells with Emily.
Raida
Today, I ran a PCR of malE31 x3, malE31delSS x2 and malE as positive control under the same PCR conditions as before, but this time with Pfu to ensure that less errors are made during the amplification. This is the last trial and I am repeating it with extra caution to avoid any kind of contamination. At the end of the day I also helped Emily with the overnight cultures.
Patrick
Still looking at protein aggregation. It seems that amyloid fibrils form after an inital nucleation step, from which other aggregate proteins begin a polymerization reaction that lengthens the fibril.
Alex
Today I set up a construction of K23900 DegP promoter with the I13507 and I13504 GFP/RFP devices. The Depg promoter was in the pSB1AK3 plasmid and was the vector which was cut using spe1 and pst1 sites. The reporter devices were cut using Xba1 and Pst1 sites. 600ng of DNA was used for each of the inserts, and 250ng of DNA was used.
 "
"