Team:Newcastle/20 August 2010
From 2010.igem.org

| |||||||||||||
| |||||||||||||
Contents |
Subtilin Immunity and rocF
Gel Electrophoresis and Gel Extraction
Aims
Following yesterday's Phusion PCR reactions, we plan to first perform gel electrophoresis of the amplified DNA sequences to check if they are of the correct sizes. If they appear to be successful we will then perform gel extraction, and finally nanodrop to record the level of DNA.
Materials and protocol
Please refer to the:
- gel electrophoresis,
- gel extraction and
- NanoDrop spectrophotometer protocols.
Results
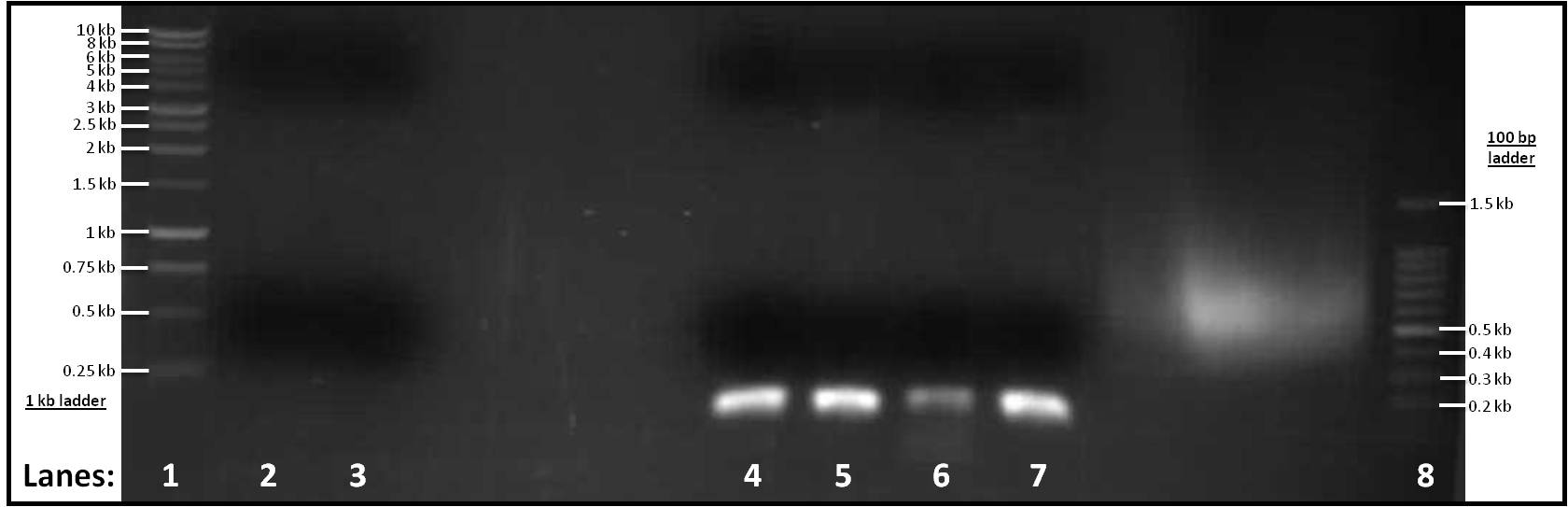
Figure 1: Gel electrophoresis of the amplified PCR products
- Lane 1: 1 Kb ladder
- Lane 2: pSB1C3 (for Subtilin Immunity)
- Lane 3: pSB1C3 (for rocF)
- Lane 4: pVeg (for Subtilin Immunity)
- Lane 5: terminator (for Subtilin Immunity)
- Lane 6: pSpacoid (for rocF)
- Lane 7: terminator (for rocF)
- Lane 8: 100 bp ladder
Discussion
After looking at the results of our gel, there are no distinct band in lanes 2-3, therefore pSB1C3 vector did not work for both Subtilin Immunity or rocF. However, there are single bands in lanes 4-7 at similar positions to each other. They are about ... in size which is what is expected.
As lanes 2-3 showed no bands, we decided to make a range of Tms to perform another set of PCR reactions to check whether the Tm was the problem. Three Tms were used: 60°C, 65°C and 70°C and six tubes were set up; a duplicate was created for each Tm. After the PCR reactions were finished, there still wasn't any visible pSBIC3 vector band showing up. The problem could have occurred due to the working stocking solution for our primers. Our next step was to re-make the working stock solution for our primers. Also, we had decided to use the Tms 55°C, 60°C, 65°C, 70°C - duplicate for each, making eight PCR tubes in total.
As lanes 4-7 had bands at the right positions, gel extraction of these four parts was carried out. The concentration of DNA was then measured using the Nanodrop spectrophotometer.
Conclusion
The PCR machines were left to run and results of the eight PCR reactions at the 4 different temperatures shall be obtained on Monday. After a test gel has been run and if bands appear at the right sizes then gel extraction will be carried out.
yneA
Gel Extraction
Aims
To purify the PCR products of digested yneA, pGFPrrnB and pSB1C3 from yesterday, as well as the promoter and double terminator for the rocF BioBrick.
Materials and Protocol
Please refer to gel extraction and nanodrop.
Results
Results of NanoDrop from gel extraction:
| DNA | Concentration (ng/µl) |
|---|---|
| yneA | 8.5 |
| pGFPrrnB | 15.3 |
| pSB1C3 | 6.4 |
Discussion
The concentration of yneA, pGFPrrnB and pSB1C3 are not very high.
Conclusion
We continue doing ligation, but also another set of minipreps so that we have a better chance of producing better results.
Miniprep
Aim
To produce stocks of yneA, pGFPrrnB and pSB1C3.
Materials and Protocol
Please refer to miniprep and nanodrop.
Results
The table below shows results of concentration (in ng/µl) from NanoDrop:
| Sample no. | yneA | pGFPrrnB | pSB1C3 |
|---|---|---|---|
| 1 | 432.5 | 238.9 | 126.1 |
| 2 | 347.6 | 230.7 | 107.6 |
| 3 | 380.2 | 390.9 | 121.5 |
| 4 | 377.9 | 236.2 | 112.8 |
Discussion
Results from the NanoDrop show concentration values of more than 100ng/µl, which means we have obtained good concentration of DNA from the miniprep.
Conclusion
We keep the miniprep products as stocks for future use.
Transformation of Ligated Products
Aims
To produce more colonies of the ligated products of yneA with pGFPrrnB and pSB1C3.
Materials and Protocol
Please refer to transformation of E. coli.
Ligation
Aims
To ligate yneA with pGFPrrnB and yneA with pSB1C3 (A repeat of yesterday).
Materials and Protocol
Please refer to ligation.
Results, Discussion and Conclusion
Please refer to 23.08.10.
Plating Bacillus subtilis BFS678
Has pMutin4 (and thus lacI) integrated into the chromosome...will be useful for characterisation.. will be transforming this strain with our filamentous cell and arginase BioBricks.
 
|
 "
"