Team:Paris Liliane Bettencourt/Project/SIP/Results
From 2010.igem.org
We have calculated SIP words for each team and each year since 2007 using SIP.C (see the code menu for more information), for each them we have took the top sip valued words. We use TOP_SIP.C to make that, this software see what is the hight SIP value and then take N words with this hight SIP.
When we got our words, we put in a [http://www.wordle.net/ software], to display them with graphical effect : words with hight frequencies has a big font, and words with low frequencies has a small font.
Just to give you a glimpse of what our software can do and how it can characterise a project, here is some examples we made on last years' winners and on the Paris 2007 team. The aim of all this is to try to extract the main ideas or key terms of a given wiki.
Contents |
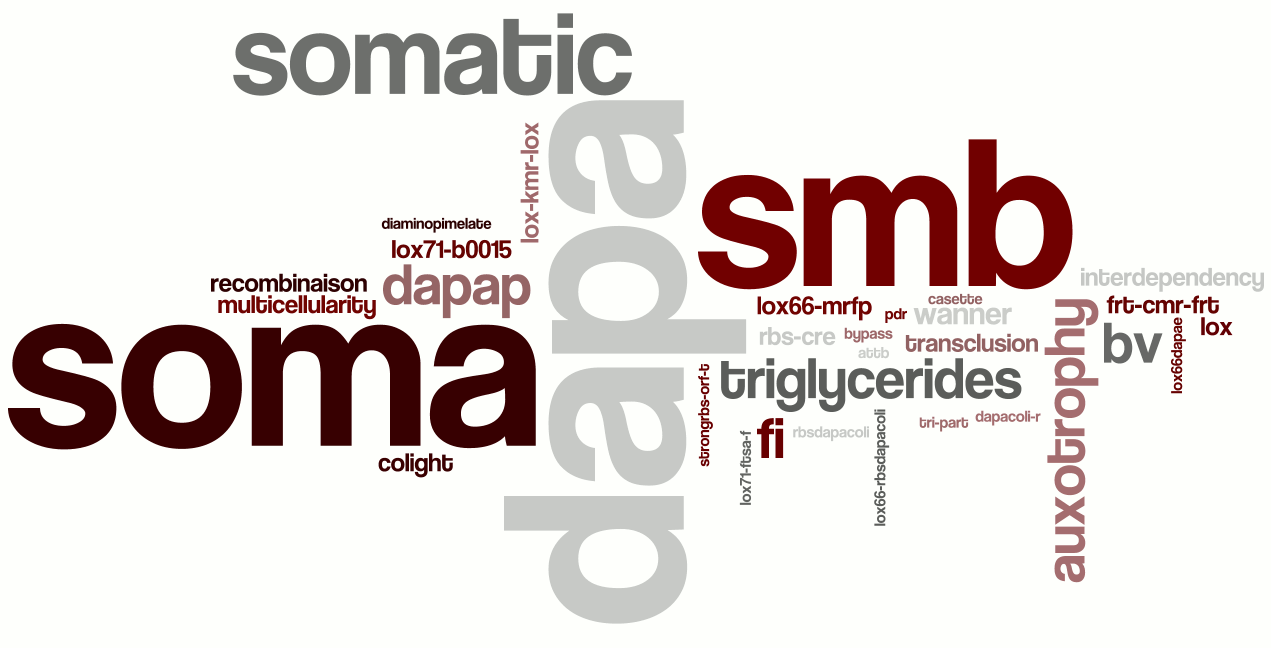
Paris Team
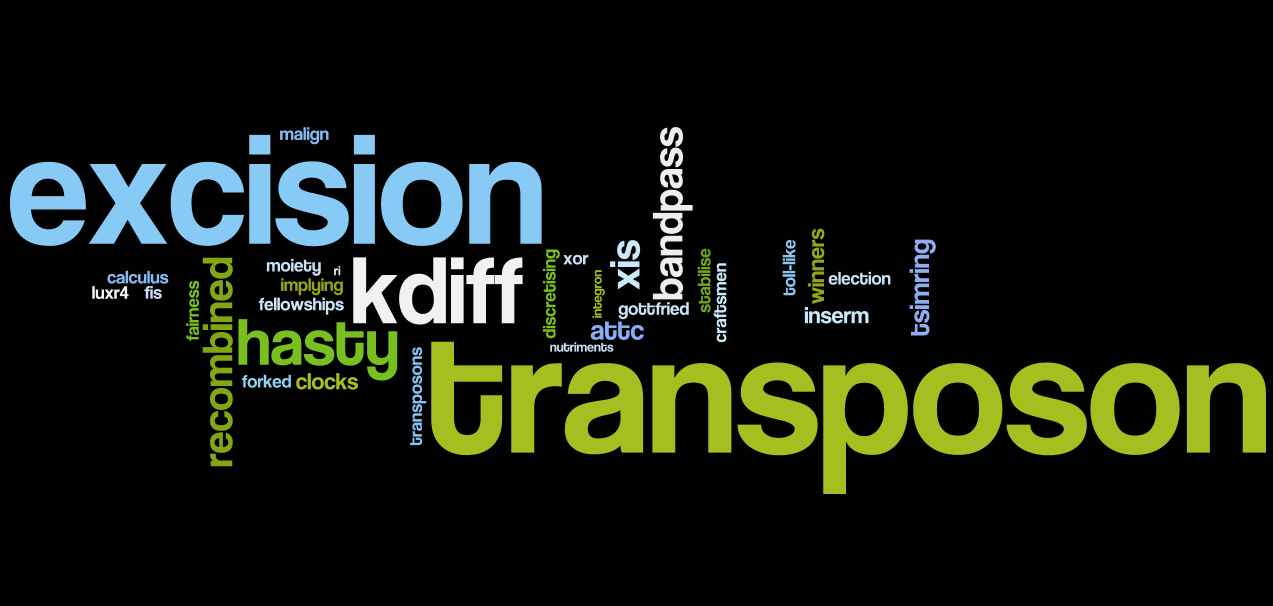
This is the analysis for [http://parts.mit.edu/igem07/index.php/Paris team Paris 2007].We have selected 60 words and filtered out all words like "p1", "f23". People name and miss-spelling words are removed to have a better display.
Just to let judge by yourself, here is the abstract of the 2007 Paris team :
"The aim of our project is to engineer the first synthetic multicellular bacterium, the SMB. This new organism is a novel tool for the engineering of complex biological systems. It consists in two interdependent cell lines. The first, dedicated to reproduction is the germ line (red cells in the simulation below). It is able to differentiate into the second line: the soma (green cells), which is sterile and dedicated to support the germ line. The germ line is auxotroph for DAP (diaminopimelate) which is provided by the soma. There is thus an interdependency relationship. The soma, being sterile, requires the germ line for its generation, while the germ line needs the soma to complement its auxotrophy. We provide here both experimental and computational evidences that this system can work, as well as the almost complete construction of the SMB."
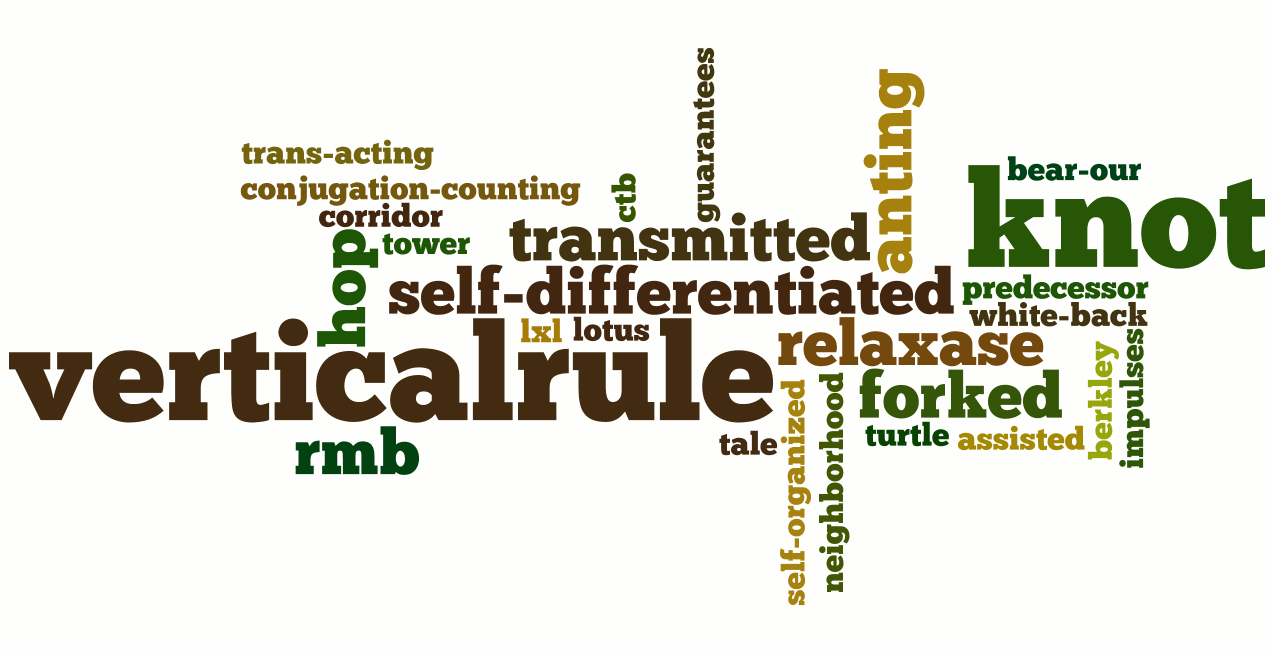
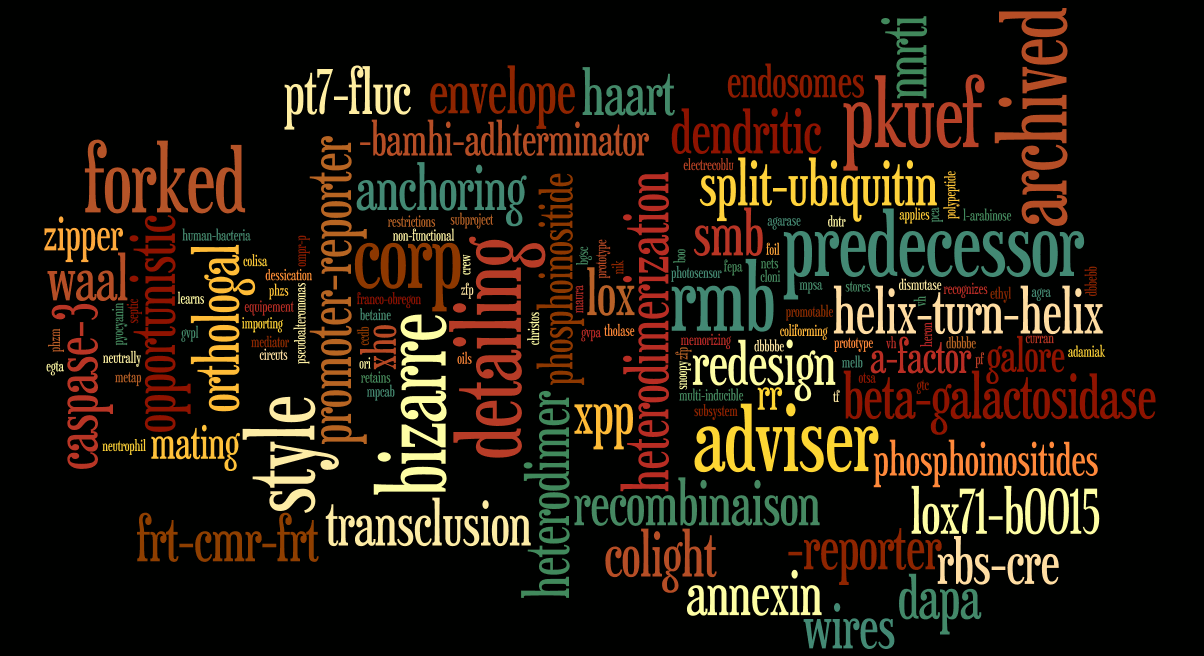
Last iGEM Winner
Here are the images for the three last iGEM winner with their related abstract. People name and miss-spelling words are removed to have a better display.
Peking 2007
"Our projects concern with the ability for bacterial cells to differentiate out of homogeneous conditions into populations with the division of labor. We aim at devices conferring host cells with the ability to form cooperating groups spontaneously and to take consecutive steps sequentially even when the genetic background and environmental inputs are identical. To break the mirror in such homogeneous condition, we need two devices respectively responsible for temporal and spatial differentiation. The implementation and application of such devices will lead to bioengineering where complex programs consisted of sequential steps (structure oriented programs) and cooperating agencies (forked instances of a single class, object and event oriented) can be embedded in a single genome. Although this "differentiation" process resemble the development of multicellular organism, we tend to use a more bioengineering style analogy: assembly line. Or maybe after some years from now, this will not be just an analogy."
Slovenia 2008
"Almost half of the world's population is infected with bacteria Helicobacter pylori, which colonize gastric mucosa, causing gastritis and ulcers and is recognized as a type I carcinogen by WHO. An effective vaccine against H. pylori is not available, although it would be a durable solution, particularly in a formulation affordable to the third world population. H. pylori evades the immune surveillance by modifying several of its components including flagellin to avoid detection by several Toll-like receptors. The goal of our project was to prepare a modular designer vaccine, using the principles of synthetic immunology. An effective vaccine has to trigger activation of adaptive immunity, which is directed against microbial proteins or polysaccharides as well as of innate immunity, which is usually achieved by the addition of adjuvants of whole microbes. We prepared a set of “immunobricks” with defined functions in activation of the immune system and can be combined to achieve a desired response. In the first approach we have modified H. pylori flagellin to be able to activate TLR5, making it »visible« to the immune system. To this chimeric flagellin we attached either complete protein or a designed a multiepitope of several virulence factors of H. pylori. We prepared three implementations of this system in the form of recombinant proteins, engineered bacteria and DNA vaccines, demonstrated responsiveness of each of them in cell culture assays, cellular localization and even obtained significant antibody response in laboratory animals only weeks after vaccination. The second approach was to extend the range of activation of innate immune response to different Toll-like receptors by linking antigen to different TLR segments, which are constitutively activated by the addition of a dimerization domain. In this case we could direct localization of resulting fusion receptors to either cell membrane or cellular vesicles, which should assist in proper antigen processing and presentation. The power of this approach is that we could mimic synergistic activation of several TLRs by pathogenic microbes, while having the advantage of safety of a defined subunit vaccine."
Cambridge 2009
"The Cambridge 2009 iGEM team has created two kits of parts that will facilitate the design and construction of biosensors in the the future.
Previous iGEM teams have focused on genetically engineering bacterial biosensors by enabling bacteria to respond to novel inputs, especially biologically significant compounds. There is an unmistakable need to also develop devices that can 1) manipulate input by changing the behaviour of the response of the input-sensitive promoter, and that can 2) report a response using clear, user-friendly outputs. The most popular output is the expression of a fluorescent protein, detectable using fluorescence microscopy. But, what if we could simply see the output with our own eyes?
We successfully characterised a set of transcriptional systems for calibrated output - Sensitivity Tuners. We also successfully expressed a spectrum of pigments in E. coli, designing a set of Colour Generators."
iGEM Ranking
To get a feeling of the major trends for winning projects and try to guess the potential winners, we determined a list of frequently "winning" SIPs. The idea is first to see what kind of projects are rewarded, then to try to see if a trend emerges by combining the last years' winners. We attribute points (coefficients) to the SIPs of the winners using the following rules :
- Grand prize : 5 pts.
- Finalist : 3pts.
- Other prize : 1pt.
- Winner : 5pts.
- 1st Runner up : 4pts.
- 2nd runner up: 3pts.
- Runner up : 2pts.
2007 year :
- Peking: 6 pts.
- Ljubljana: 4 pts.
- Paris: 4 pts.
- UCSF: 3 pts.
- USTC: 4 pts.
- Glasgow: 1 pt.
- Alberta: 1 pt.
- Cambridge: 1 pt.
- Melbourne: 1 pt.
- UC Berkeley: 1 pt.
- Calgary: 1 pt.
- ETHZ: 1 pt.
2009 year
- Alberta: 1 pt.
- ArtScienceBangalore: 1 pt.
- BCCS-Bristol: 1 pt.
- Berkeley Software: 1 pt.
- Cambridge: 6 pts.
- EPF-Lausanne: 1 pt.
- Freiburg bioware: 4 pts.
- Groningen: 2 pts.
- Heidelberg: 6 pts
- Illinois-Tools: 1 pt.
- Imperial College London: 4 pts.
- Paris: 1 pt.
- Stanford: 1 pt.
- TUDelft: 1 pt.
- ULB-Brussels: 1 pt.
- UNIPV-Pavia: 1 pt.
- Valencia: 5 pts.
All high ranked team (2007-2008-2009). We've take 8 words for each.
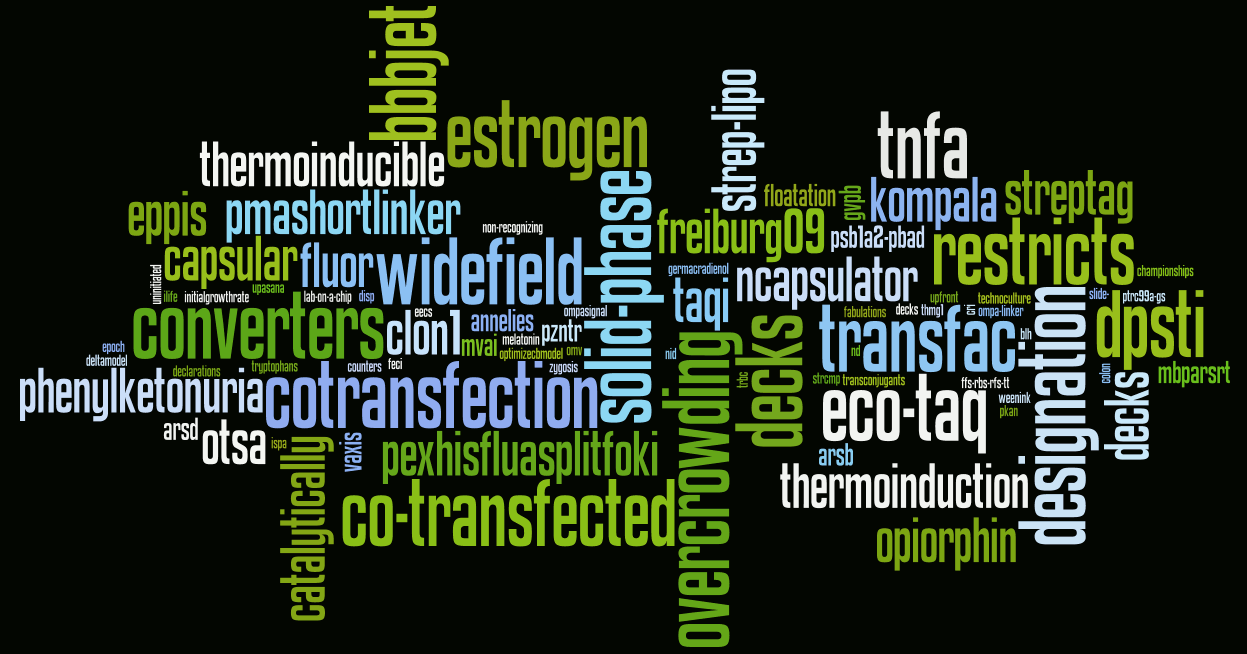
Paris 2010
And last, we have try to see your sip wiki, we have use the 2009 dictionary to make that.
 "
"