Team:Paris Liliane Bettencourt/Brainstorming
From 2010.igem.org
Brainstorming
I. Band-pass filter
What is a band-pass filter ?
Like a lot of genetic circuits, band-pass filter is coming from electronic. In electronic the definition of a band-pass filter is the Combination of a high-pass filter and a low- pass filter in series.
For our project, it means that we have a system which permit the expression of a gene (let’s say GFP) only in a range of concentration ([f1,f2] in the fig 1).
Background
It already exists few bandpass filter (thanks to ribozyme for example) but the most interesting one is an externally tunable bandpass filter using enzyme activity and a molecular inducer to shift the frame of expression.
In the presence of Tet and Amp, growth only in narrow concentration range of Amp. Range of Amp controlled by enzyme activity means IPTG.
What kind of applications?
For the moment, bandpass filter are mostly used for spatial patterning which permit to study cellular differentiation
We can use a bandpass filter has a band pass detector which means to couple the bandpass filter to a biosensor and express a reporter gene only in a range of concentration. But it will be not as fast as a chemical diagnostic technics for example.
If we can manage to create a modulable bandpass filter it could be usefull for metabolic engineering. If the inducer is a metabollite in a bottleneck reaction or a toxic metabollit, we can find ,measuring the concentration of the protein of interest, for which concentration of metabolite (means for which bandpass filter) the pathway is optimum.
What kind of circuits can we use ?
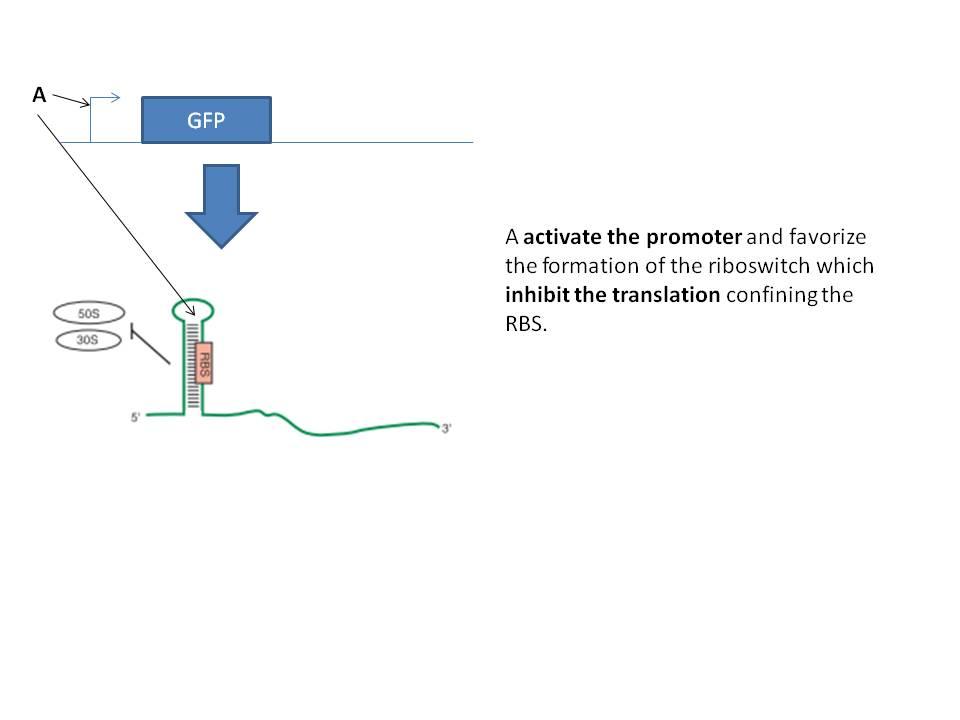
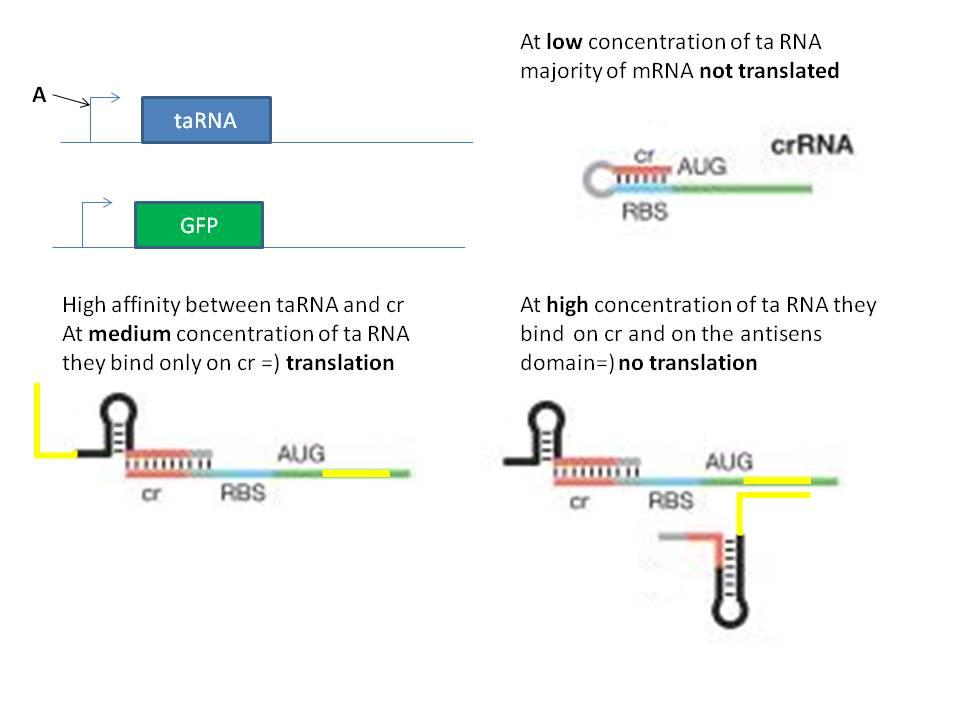
The first idea was the ciruit below. The main idea is to tune the strength of the promoter and the riboswitch to get a bandpass filter.
The second idea was to implement in a single RNA molecule the inhibition and activation part necessary to the bandpass filter. The activation part seems to be "well known" but implement an antisens domain has not been done because the mechanism of this kind of stuff is not well known. We can try to do it, it will be interesting to reate a way to induce the degradation of a mRNA with a natural system.
In conclusion I think the part we have to develop if we want to do this project is a nice application, maybe coupled to the anti-toxin/toxin characterization project it will be cool.
References
- [http://www.ncbi.nlm.nih.gov.gate2.inist.fr/pmc/articles/PMC2700907/ An externally tunable bacterial band-pass filter]
- Engineered riboregulators enable post-transcriptional control of gene expression
II. Bacterial calculator
General idea
Implement logic gates that could be used in combinations in a microfluidic device to realize simple digital computations (addition and substraction).
How it would work
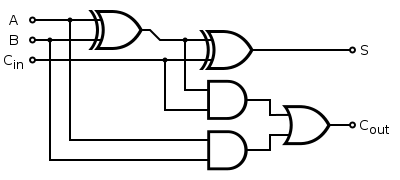
- A full 1bit adder consists of 2AND, 2XOR and 1OR gate :
- The design of a modular AND gate is shown in [http://www.ncbi.nlm.nih.gov/pubmed/17700541 Environmental signal integration by a modular AND gate] (J Christopher Anderson, Christopher A Voigt, and Adam P Arkin, 2007).
- The design of XOR and OR gates is to be made.
- The design is to be implemented on a microfluidic device to prevent cells from contaminating adjacent chambers, and to control the moment of the signal transmission from one chamber to the next one.
- There would be a different cell type for each gate. The adder would be constructed on a microfluidic device where each chamber contains 1 cell type and the chambers are connected according to the circuit design. As soon as 1 full adder is constructed, it can be connected to its replica, providing a 2bit adder etc.
Advantages of the project
- Show that cells are able to integrate signals, interact with other cell types and thus work as transistors, realizing digital computations.
- Microfluidic devices are interesting tools to use in the project.
- The AND gate was already used by another iGEM team, there are already some BioBricks and we know this kind of tuning is feasible for an iGEM project.
Drawbacks and difficulties
- Will we be able to construct modular OR and XOR gates?
- What would be the application of such a circuit?
- Working with a microfluidic device could be difficult in a way to control the chamber connections (valves opening and closing the channels).
References
- [http://www.play-hookey.com/digital/adder.html How a binary adder works]
- [http://www.ncbi.nlm.nih.gov/pubmed/17700541 Design of a modular AND gate]
- Team that used this AND gate design for their project
- Team that used microfluidic devices, but without much success
III. RepRap project
General idea
Use the idea of the RepRap machine that can "reproduce itself" and go further, engineering bacteria that would produce plastic for the RepRap machine and then use it to print Petri dishes, pipettes etc. to grow new bacteria. Also, make coloured plastic or plastic with new characteristics.
How it would work
- Bacteria have already been engineered to produce plastic (PLA and its PHA copolymers) in large quqntities, so that it forms agregates inside the cells until they burst. Our work would be to recreate this circuit with biobricks.
- Until now, the RepRap machine prints with the standart white plastic (PLA for example). It melts PLA at about 180-190°C, so the dye must be stable at this temperature. It seems that the current dyes used for PLA (not in RepRap) are in fact PET dyes and not really specific to PLA. [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B8G3X-4PGPM55-W&_user=658968&_coverDate=09%2F30%2F2007&_rdoc=1&_fmt=high&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000035758&_version=1&_urlVersion=0&_userid=658968&md5=85c3a1aff17c40df2c3ce07eb57d3e98 This article] states that anthraquinone dyes are more specific of PLA.
Advantages of the project
- RepRap is a highly interesting project and there are people in Paris who are working on it.
- This project would have a direct application.
Drawbacks and difficulties
- How should we deal with the rights of the article authors? RepRap is an opensource project, but the authors will probably want to get money from their plastic-producing bacteria and won't cooperate with us so easily.
- Making bacteria produce colours is quite a dead-end : it should be produced in a sufficient quantity and remain stable at high temperatures. Producing anthraquinone dyes by modifying metabolic pathways in E.coli seems too difficult.
- We should also be able to separate the bacteria from the dye. Should we grow both types of bacteria together or separately?
References
- [http://www.ncbi.nlm.nih.gov/pubmed/19937726?dopt=AbstractPlus Biosynthesis of Polylactic Acid and Its Copolymers Using Evolved Propionate CoA Transferase] and [http://www.ncbi.nlm.nih.gov/pubmed/19937727 PHA Synthase and Metabolic Engineering of Escherichia coli for theProduction of Polylactic Acid and Its Copolymers] the two articles by the same authors on the production of PLA and its copolymers
- [http://reprap.org/wiki/Main_Page RepRap website]
- [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B8G3X-4PGPM55-W&_user=658968&_coverDate=09%2F30%2F2007&_rdoc=1&_fmt=high&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000035758&_version=1&_urlVersion=0&_userid=658968&md5=85c3a1aff17c40df2c3ce07eb57d3e98 Dyes with high affinity for polylactide], article about new dyes based ou anthraquinone
IV. Morse code
General idea
Making bacteria "speak" with morse code, i.e. emit signals that would be different not only in color (GFP, RFP, YFP, CFP), but also in length (short/long pulse).
How it would work
- The initial idea is represented here :
The cell coding for the 1st letter of the word is induced by an endogenous molecule (here IPTG). Downstream of the promoter are 3 genes:
- GFP with an ssrA sequence in the end, so that it would be recognized by a protease and rapidly destroyed - like in 10 minutes (green line at the bottom image). It's the “long” signal.
- A1, the activator (or the enzyme that produces the activator molecule) for the promoter of the 2nd construct . A1 would have a longer life and not be destroyed so rapidly (yellow line)
- R1 with an ssrA sequence, so that it would be destroyed as fast as the GFP. R1 would inhibit A1. In this way, A1 will be able to work as an activator of P1 only after R1 (and GFP) are destroyed and assure that the 2nd signal starts only after the first one ends. An example of A1 and R1 I can think of is A1=AraC and R1=enzyme that cleaves arabinose.
When the first signal is finished, A1 activates P1 and a new triad of proteins is expressed. This time GFP has another sequence at the end so that it's degraded in less time, like 3 minutes. It provides the “short” signal. A2 and R2 would need to work according to the same principle as A1 and R1 to activate P2 after the signal is finished. Downstream of P2 is another GFP for “long” signal, and LuxI. As letter K is coded by 3 signals '_ . _' after the third signal is emitted, the cell has finished its message and has to induce another cell type coding for the second letter. The construct in the second cell would be the same, except that the first promoter will now be LuxR, and the sequence of the signals will be different. The second cell type could give the response in RFP to avoid confusion with noise from the first cell type. The activation of the second cell type being performed by AHL, it could synchronize 2nd cell type population to start the 2nd letter transmission together.
The third letter would be activated in the same way, only by another AHL molecule.
- This idea was further discussed and another way to produce pulses of different length was proposed, as described in the article [http://www.ncbi.nlm.nih.gov/pubmed/15096621 Spatiotemporal control of gene expression with pulse-generating networks] (Subhayu Basu, Rishabh Mehreja, Stephan Thiberge, Ming-Tang Chen, and Ron Weiss, 2004) :
Advantages of the project and new ideas
- Making a larger "library" of visible cell responses (combinations of lights and pulse lengths).
- Is there a way to make a "frequency filter"? For example, to control the translocation of the transcription factors into the nucleus.
Drawbacks and difficulties
- Talking with Morse code isn't very interesting, bacteria can do much better than simply long and short signals.
- If we want a longer word, we would need more different AHL molecules and LuxRs. Also, different cells for the same letter of alphabet if it appears in the word twice.
- What would be the application? Emitting a predetermined signal isn't useful. Maybe the pattern of the emitted signals could change according to the inputs?
References
- [http://www.ncbi.nlm.nih.gov/pubmed/15096621 Article about pulse generating networks]
V. Measuring the strength of gene expression by an internal control
An antitoxin controlled by a calibrated promoter/riboswitch circuit and its toxin controlled by an unknown rbs/promoter. Culture in a 96-well plate with two gradients of inducer, use the concentrations under which the culture grows to back calculate the strength of the unknown rbs/promoter.
VI. Storage of organophosphorus compounds
There exists some enzymes which cleave organophosphorous and it would be interesting to inactivate the cleavage activity but keep and improve the binding activity to permit the storage of organophosphorous in the cell. In water treatment they use sedimentation so if we can permit a fast sedimentation of the cells containing organophosphorous we can have rich mud for agriculture.
VII. Programmed cell death with delay
The idea of the cell death program with delay is to be able to release engineered bacteria safely in the environment.
For the cell death program it has to :
- lyse DNA (production of nuclease)
- be fast to reduce the possibility of mutants
- maybe target multiple independent elements to avoid mutants
To produce the delay :
- an input which triggers the slow production of an inducer for the cell death program. If it’s sufficiently slow and the induction with a high threshold there will be a delay.
- put the gene for the inducer of the death cell program behind a promoter active just after the cellular division or related to a cell cycle. If the half life of the molecule is sufficiently high there will have some accumulation with division. Like this we can count the number of division between the death.
For the KO of a self-maintenance gene, maybe it will have some deleterious effect on the activity of the cell and so on the interesting action implemented.
We need to limit as much as possible horizontal transfers that could disrupt your system.
VIII. Control of biofilm production/degradation
It could be interesting to be able to control the biofilm production in response to an external agent.
-Control the start of the production
-Be able to destroy it on demand
IX. Others
Bacteria making sound, Control of bacteria’s movements, Live painting using bacteria, Antifouling, Random number generator, Cycle Phase Detector using fluorescent markers, Self-regenerating polymer, Games (Bacterial soccer, Labyrinthe, Otello, Morpion, Football, Ecowar), Creation of a bacterial society (auto-organisation with different cell types which need each other to live)...
 "
"