Team:Tokyo Tech/Project/wolf coli/lacIM1
From 2010.igem.org
(→Abstract) |
|||
| Line 49: | Line 49: | ||
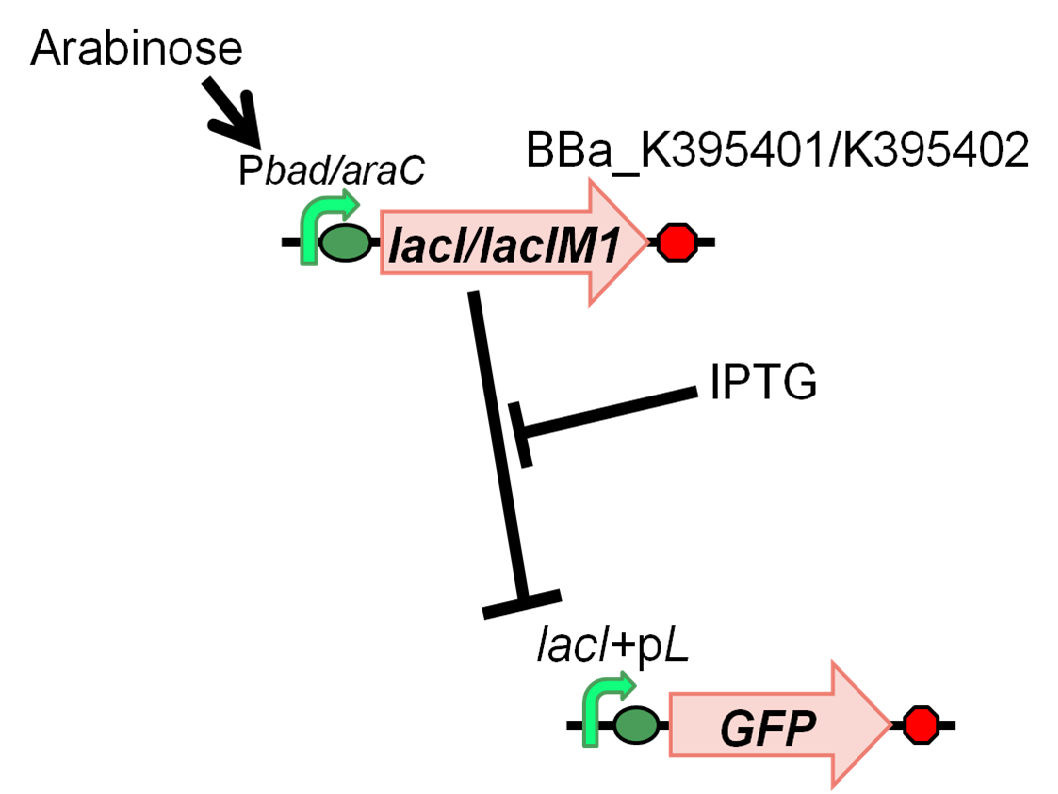
[[Image:Tokyotech LacIM1 system ver4.png|left|thumb|300px|Figure4-2-1. ]] | [[Image:Tokyotech LacIM1 system ver4.png|left|thumb|300px|Figure4-2-1. ]] | ||
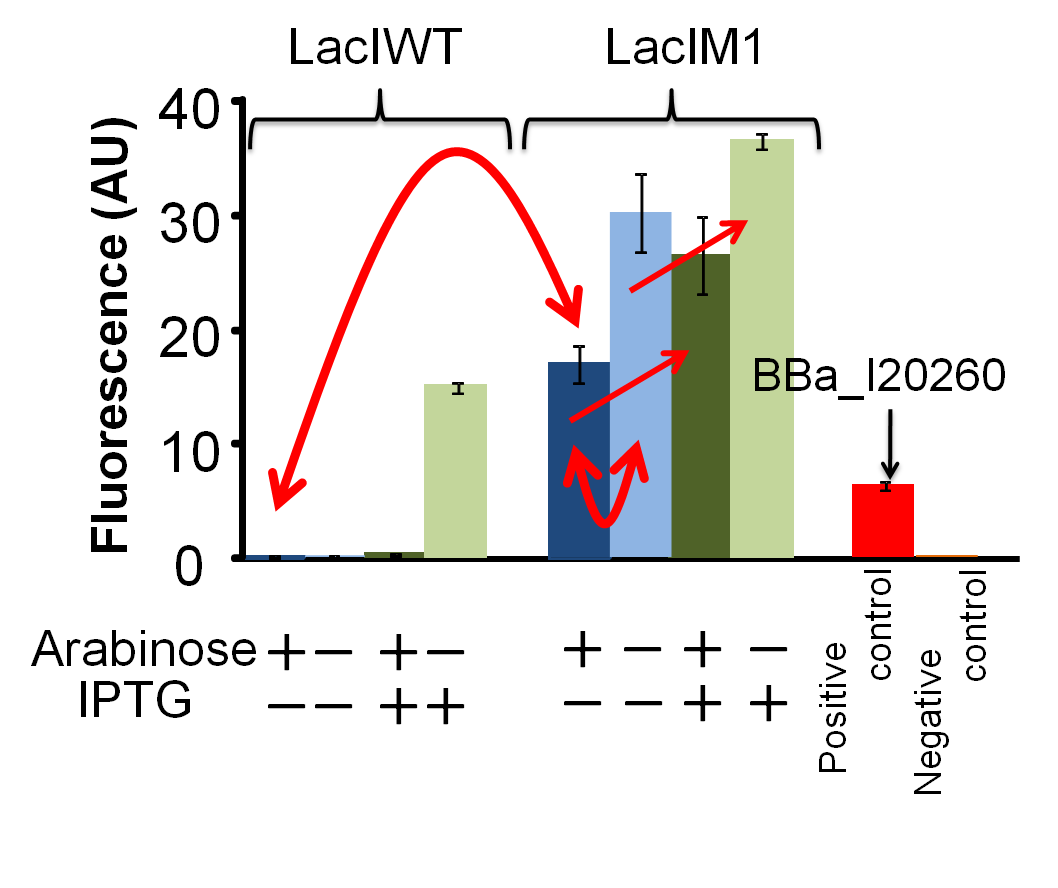
[[Image:Tokyotech_LacIM1_data_ver2.png|right|thumb|300px|Figure4-2-2. Repression efficiency of LacIM1 (BBa_K395401) / LacIWT (BBa_K395402) exposed to arabinose and IPTG. This work is done by Mitsuhiko Odera ]] | [[Image:Tokyotech_LacIM1_data_ver2.png|right|thumb|300px|Figure4-2-2. Repression efficiency of LacIM1 (BBa_K395401) / LacIWT (BBa_K395402) exposed to arabinose and IPTG. This work is done by Mitsuhiko Odera ]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
==Introduction== | ==Introduction== | ||
Revision as of 19:13, 27 October 2010
4-2 lacIM1 for band-detect network
Contents |
Abstract
We characterized LacIM1 (BBa_K082026), a mutation of LacIWT,which is a key component in the band-detect network. In fact, this part was registered by USTC(2008) [3], however, sequence data of BBa_K082026 is incorrect and it was not well characterized in the BioBrick registry. Therefore, we registered this part as BBa_K395400 and confirmed that product of lacIM1 shows weaker repression to Plac than its wild type. In order to measure the function of lacI proteins, we constructed following two plasmids, BBa_K395401 (LacIM1) and BBa_K395402 (LacIWT), which have an arabinose inducible promoter. We measured GFP expression dependent on the input of arabinose and IPTG(Fig. 4-2-1).
Introduction
What is LacIM1?
As I wrote before, LacIM1 shows weaker repression compare to LacIWT because of its lower affinity to lac promoter [1].
Requirement for band-detect network
The band-detect network exhibits transient gene expression in response to concentration of chemical signals. In the band-detect network, LacIM1 is a key component because its repression efficiency can afford the network the desired non-monotonic response.
Results
(Figure4-2-2.
Repression efficiency of LacIM1 ([http://partsregistry.org/Part:BBa_K395401 BBa_K395401]) / LacIWT ([http://partsregistry.org/Part:BBa_K395402 BBa_K395402]) exposed to arabinose and IPTG.
Dark-blue bars, 0.2% arabinose and 0 mM IPTG;
blue bars, 0% arabinose and 0 mM IPTG;
dark-green bars, 0.2% arabinose and 1 mM IPTG;
green bars, 0% arabinose and 1 mM IPTG;
red bar, positive control
orange bar, negative control
GFP expression is dependent on the input of arabinose and IPTG. After 4 hours of arabinose and IPTG induction. Cultures were analysed by fluorometer (FLA5000、FUJI).
As positive control, we used BBa_I20260, which is measurement kit of the iGEM.)
After 4 hours of arabinose and IPTG induction. Cultures were analysed by fluorometer (FLA5000, FUJI).
This is the result of LacIM1.
In the absence of both arabinose and IPTG, GFP expression is about 1.8-fold stronger than that of the presence of arabinose and the absence of IPTG. We confirmed the dependency of GFP expression on product of lacIM1 by arabinose induction. In other words, the fact that LacIM1 represses the GFP expression was corroborated, based on our results. Moreover, Increase of GFP production by addition of IPTG, with or without arabinose induction, again shows that the repressions are dependent on LacIM1. Without arabinose induction, increase of GFP production by addition of IPTG, can be explained due to the leaky expression of LacIM1. This leaky expression could also explain the results for LacWT.
This is the result of LacIWT.
・In the presence of arabinose and the absence of IPTG, GFP expression did not occur. This results show that LacI protein was expressed by arabinose induction and could fully repressed GFP expression.
Absence of both arabinose and IPTG. Normally without arabinose induction, the GFP is supposed to be expressed. However, GFP expression did not occur. This result shows that the leaky expression of LacI protein is been sufficient for repressing GFP expression.
・Presence of both arabinose and IPTG. Normally, although arabinose induces the overproduction of LacI protein and represses the GFP expression, LacI protein can be inhibited by IPTG. Therefore, we expected the occurrence of GFP expression. However, there is no expression of GFP. This result shows that the amount of IPTG was not sufficient to inhibit the repression by overproduced LacI proteins.
・In the absence of arabinose and the presence of IPTG, as I mentioned before, leaky expression of LacI protein occurs. However, when there is only small amount of LacI protein, addition of IPTG can fully inhibit the repression of LacI protein. Thus, GFP expression occurs.
Conclusion
We confirmed that lacIM1 shows weaker repression compare to LacIWT.
Materials&Methods
Construction of E.coli strain DH5α
In order to characterize K082026(K395400), LacI Mutant, we constructed BBa_K395401 combining I0500 and K082026(K395400) on pSB1A3. and used BBa_I20260 as a positive control and used promoterless gfp on pSB3K3 as a negative control. Furthermore, we constructed BBa_K395402 combining I0500 and BBa_I 732820 on pSB1A3 as a control experiment. As a GFP reporter, we used BBa_I7106 on pSB3K3. In order to measure the function of lacI proteins, we introduced BBa_I7106 (lacI+pL-rbs-GFP-ter) on pSB3K3 and BBa_K395401/BBa_K395402 into DH5α.
Report assay
Overnight cultures of reporter strains grown at 37 °C in LB Medium containing appropriated antibiotics were diluted at least 1:100 in the medium and were incubated at 37 °C as fresh cultures. After their OD590 reached 0.3, added appropriate inducers that 1mM IPTG, and/or 0.2% arabinose (final concentration). After 4 hours of induction, 150 l of each sample was applied to 96-well plate and its fluorescence intensity was measured with a fluorometer by BPB-sp filter, which emit LB. The fluorescence intensity was calculated by dividing the measured raw fluorescence intensity by the adjusted OD590.
Refference
[1]. Basu S, Gerchman Y, Collins CH, et al. A synthetic multicellular system for programmed pattern formation. NATURE 2005;434,1130-1134
[2]. USTC(2008)
 "
"