Team:Tokyo Tech/Project/Artificial Cooperation System/Cm assay
From 2010.igem.org
| (27 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
<table id="table-01"> | <table id="table-01"> | ||
<tr> | <tr> | ||
| - | <td>[[Team:Tokyo_Tech|1 | + | <td>[[Team:Tokyo_Tech|1 Graphic abstract]]<br> |
</td> | </td> | ||
</tr> | </tr> | ||
<td>2 Apple reporter<br> | <td>2 Apple reporter<br> | ||
:[[Team:Tokyo_Tech/Project/Apple_Reporter|2-1 Color]] | :[[Team:Tokyo_Tech/Project/Apple_Reporter|2-1 Color]] | ||
| - | |||
:[[Team:Tokyo_Tech/Project/Apple_Reporter2|2-2 Fragrance]] | :[[Team:Tokyo_Tech/Project/Apple_Reporter2|2-2 Fragrance]] | ||
</td> | </td> | ||
| Line 19: | Line 18: | ||
<th>[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System|3 Artificial Cooperation System]]<br> | <th>[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System|3 Artificial Cooperation System]]<br> | ||
:[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System/lux_act_rep|3-1 lux activation/repression promoter]] | :[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System/lux_act_rep|3-1 lux activation/repression promoter]] | ||
| - | :3-2 resistance gene activation device - | + | :3-2 resistance gene activation device -YOU ARE HERE!- |
:[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System/luxI_assay|3-3 ''lux''I Assay]] | :[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System/luxI_assay|3-3 ''lux''I Assay]] | ||
:[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System/modeling|3-4 modeling]] | :[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System/modeling|3-4 modeling]] | ||
| Line 26: | Line 25: | ||
<tr> | <tr> | ||
<td>[[Team:Tokyo_Tech/Project/wolf_coli|4 wolfcoli overview]]<br> | <td>[[Team:Tokyo_Tech/Project/wolf_coli|4 wolfcoli overview]]<br> | ||
| - | :[[Team:Tokyo_Tech/Project/wolf_coli/New_Series_of_PompC|4-1 | + | :[[Team:Tokyo_Tech/Project/wolf_coli/New_Series_of_PompC|4-1 New seriesof P''ompC'']] |
:[[Team:Tokyo_Tech/Project/wolf_coli/lacIM1|4-2 lacIM1 for band-detect network ]] | :[[Team:Tokyo_Tech/Project/wolf_coli/lacIM1|4-2 lacIM1 for band-detect network ]] | ||
| - | :[[Team:Tokyo_Tech/Project/wolf_coli/System|4-3 | + | :[[Team:Tokyo_Tech/Project/wolf_coli/System|4-3 Wolf coli system]] |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 40: | Line 39: | ||
<font size="5"><b>3-2 resistance gene activation device</b></font> | <font size="5"><b>3-2 resistance gene activation device</b></font> | ||
==Abstract== | ==Abstract== | ||
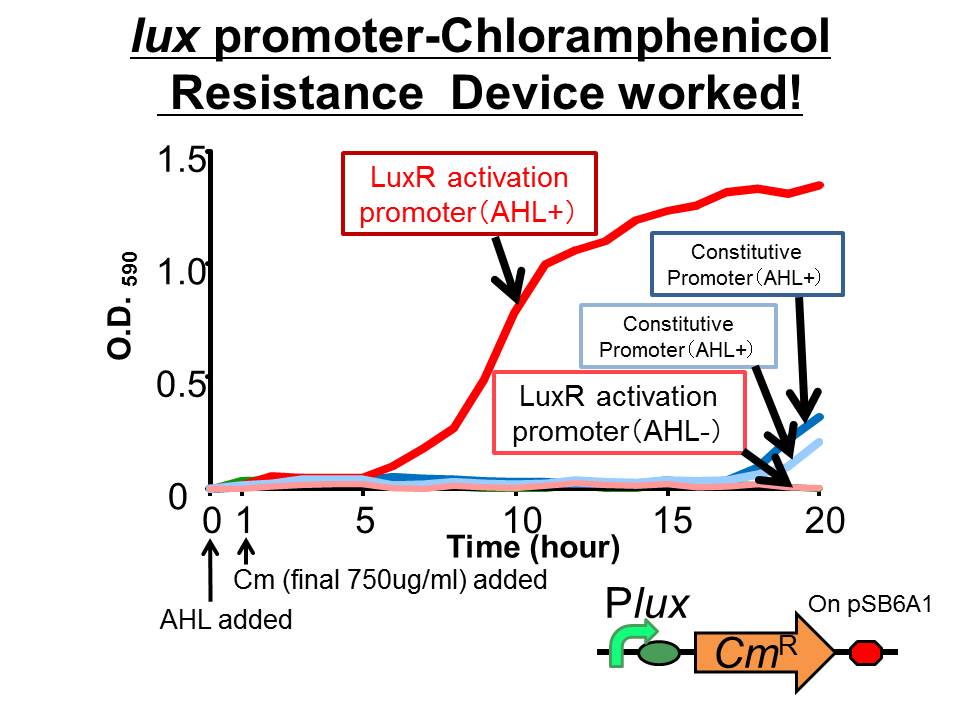
| - | We succeeded in constructing and characterizing a NEW Biobrick device | + | We succeeded in constructing and characterizing a NEW Biobrick device ( [http://partsregistry.org/Part:BBa_K395162 BBa_K395162] ). This device is a ''lux'' promoter-activated chloramphenicol resistance (''CmR''). We found that the device is activated by LuxR/3OC6HSL complex and when the device was introduced into a cell, it was able to survive even in high chloramphenicol concentration (750 ug/ml) in the presence of 3OC6HSL. |
| - | + | [[Image:tokyotech_Cm-survival.jpg|thumb|fig.3-3-1|400px|left]] | |
| - | + | ||
| - | + | ||
| - | [[Image:tokyotech_Cm-survival.jpg|thumb|400px|left]] | + | |
| Line 71: | Line 67: | ||
| + | |||
| + | ==Introduction== | ||
| + | Chloramphenicol is a well known antibiotic that is effective against a wide variety of Gram-positive and Gram-negative bacteria including ''E. coli''. Chloramphenicol stops bacterial growth because it is a protein synthesis inhibitor, which prevents peptide bond formation. When chloramphenicol is added, the synthesis of proteins is inhibited in bacteria which don't express chloramphenicol resistance gene. Then their growth stops and the number of the cell is going to decrease. However, bacteria which express chloramphenicol resistance gene are able to survive in the existence of chloramphenicol. These characteristics indicates that chloramphenicol and chloramphenicol resistance gene can be used for population control which is important to construct the Artificial Cooperation System | ||
==Results== | ==Results== | ||
| - | *The results of 20 hours incubation after addition of chloramphenicol shows that P''lux''-CmR grew when 3OC6HSL is added ,while P''lux''-CmR in the absence of 3OC6HSL was not able to grow. | + | *The results of 20 hours incubation after addition of chloramphenicol shows that P''lux''-CmR grew when 3OC6HSL is added , while P''lux''-CmR in the absence of 3OC6HSL was not able to grow. |
| - | + | On the other hand, The cells introduced the promoterless device were not able to grow independently of 3OC6HSL. In the case of the device that had constitutive promoter, the growth of the cells introduced the device were quite slow compared with that of P''lux''. However, there was no significant difference between in the presence and the absence of 3OC6HSL. | |
| - | + | <br> | |
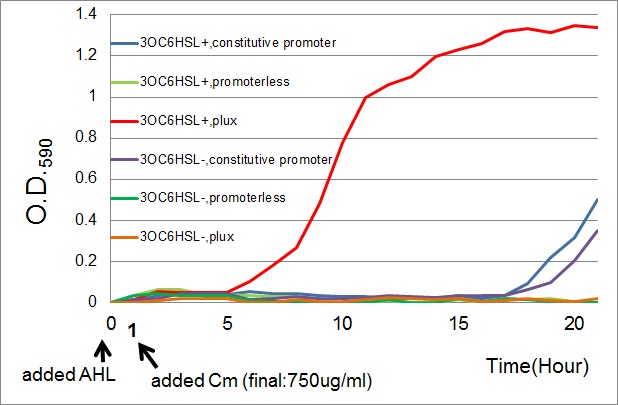
| - | [[Image: | + | [[Image:Tokyotech_cm_assay.jpg|600px|thumb|Fig.3-3-2 Growth curve of the cell introduced PlacI<sup>q</sup>-''CmR'' (BBa_K395165,blue curves), P''lux''-''CmR'' (BBa_K395162,Red curves), or promoterless-''CmR'' (BBa_K395160,green curves) with or without AHL (3OC6HSL).<br>This work is done by Yusuke Kaneta.]] |
==Conclusion== | ==Conclusion== | ||
| - | From the results, when chloramphenicol was added, the cells | + | From the results, when chloramphenicol was added, only the cells P''lux''-''CmR'' being introduced into them were able to grow because LuxR/3OC6HSL complex activated P''lux''. This means we found the condition that integrants were able to survive only in the presence of 3OC6HSL. |
==Material and Method== | ==Material and Method== | ||
| - | === | + | ===Construction P''lux''-''CmR'' on pSB6A1=== |
| - | + | RBS-''CmR'' part was cut from BBa_P1004 by PCR and inserted into pSB6A1. Next RBS-''CmR'' on pSB6A1 and P''lux'' (BBa_R0062) were cut at EcoRI /Xba sites and EcoRI/SpeI sites respectively. Then we ligated these two parts. | |
===Construction of ''E. coli'' strain DH5α=== | ===Construction of ''E. coli'' strain DH5α=== | ||
Competent cells with ''luxR'' gene on pSB3K3 were constructed. Afterward, P''lux''-''CmR'' on pSB6A1 was introduced into the cells. | Competent cells with ''luxR'' gene on pSB3K3 were constructed. Afterward, P''lux''-''CmR'' on pSB6A1 was introduced into the cells. | ||
| Line 90: | Line 89: | ||
In order to follow up the growth of the cells which P''lux''-''CmR'' was introduced into, the change of O.D. (590nm) in both the presence and the absence of 3OC6HSL was measured.<br> | In order to follow up the growth of the cells which P''lux''-''CmR'' was introduced into, the change of O.D. (590nm) in both the presence and the absence of 3OC6HSL was measured.<br> | ||
(samples)<br> | (samples)<br> | ||
| - | #P''lux''-''CmR''[http://partsregistry.org/Part:BBa_K395162 BBa_K395162] | + | #P''lux''-''CmR ''[http://partsregistry.org/Part:BBa_K395162 BBa_K395162] |
| - | #constitutive promoter (PlacI<sup>q</sup>)-''CmR'' (positive control)[http://partsregistry.org/Part:BBa_K395165 BBa_K395165] | + | #constitutive promoter (PlacI<sup>q</sup>)-''CmR '' (positive control)[http://partsregistry.org/Part:BBa_K395165 BBa_K395165] |
| - | #promoterless-''CmR'' (negative control)[http://partsregistry.org/Part:BBa_K395160 BBa_K395160]<br> | + | #promoterless-''CmR '' (negative control)[http://partsregistry.org/Part:BBa_K395160 BBa_K395160]<br> |
| - | LB medium | + | LB medium was used as liquid culture. Antibiotics (ampicillin (Amp), kanamycin (Kan), and chloramphenicol (Cm)) were dissolved in distillation water and stored in 25 mg/ml concentration; their final concentration in the medium was adjusted to 50ug/ml, 30ug/ml, and 750ug/ml respectively. |
| - | #The seed cultures of the samples were inoculated from glycerol stock solution and grown separately overnight at 37°C in LB medium containing Amp&Kan. | + | #The seed cultures of the samples were inoculated from glycerol stock solution and grown separately overnight at 37°C in LB medium containing Amp & Kan. |
| - | #Then, we diluted the cultures 100 | + | #Then, we diluted the cultures to 100-fold by two types of 3 ml of fresh medium. One type was the medium containing 3OC6HSL (final concentration:100nM). Another medium contained DMSO as experiment control. Both of the cultures were incubated at 37°C |
| - | # | + | #1 hour after dilution, each culture was separated into two new tubes. Next, 1.5 ml LB medium each containing Cm (final concentration:750ug/ml), Amp, Kan,3OC6HSL or DMSO was added to each tube. |
| - | # | + | #O.D.590 was measured every hour, up to 20 hours from dilution. |
==Reference== | ==Reference== | ||
Latest revision as of 03:46, 28 October 2010
3-2 resistance gene activation device
Contents |
Abstract
We succeeded in constructing and characterizing a NEW Biobrick device ( [http://partsregistry.org/Part:BBa_K395162 BBa_K395162] ). This device is a lux promoter-activated chloramphenicol resistance (CmR). We found that the device is activated by LuxR/3OC6HSL complex and when the device was introduced into a cell, it was able to survive even in high chloramphenicol concentration (750 ug/ml) in the presence of 3OC6HSL.
Introduction
Chloramphenicol is a well known antibiotic that is effective against a wide variety of Gram-positive and Gram-negative bacteria including E. coli. Chloramphenicol stops bacterial growth because it is a protein synthesis inhibitor, which prevents peptide bond formation. When chloramphenicol is added, the synthesis of proteins is inhibited in bacteria which don't express chloramphenicol resistance gene. Then their growth stops and the number of the cell is going to decrease. However, bacteria which express chloramphenicol resistance gene are able to survive in the existence of chloramphenicol. These characteristics indicates that chloramphenicol and chloramphenicol resistance gene can be used for population control which is important to construct the Artificial Cooperation System
Results
- The results of 20 hours incubation after addition of chloramphenicol shows that Plux-CmR grew when 3OC6HSL is added , while Plux-CmR in the absence of 3OC6HSL was not able to grow.
On the other hand, The cells introduced the promoterless device were not able to grow independently of 3OC6HSL. In the case of the device that had constitutive promoter, the growth of the cells introduced the device were quite slow compared with that of Plux. However, there was no significant difference between in the presence and the absence of 3OC6HSL.
Conclusion
From the results, when chloramphenicol was added, only the cells Plux-CmR being introduced into them were able to grow because LuxR/3OC6HSL complex activated Plux. This means we found the condition that integrants were able to survive only in the presence of 3OC6HSL.
Material and Method
Construction Plux-CmR on pSB6A1
RBS-CmR part was cut from BBa_P1004 by PCR and inserted into pSB6A1. Next RBS-CmR on pSB6A1 and Plux (BBa_R0062) were cut at EcoRI /Xba sites and EcoRI/SpeI sites respectively. Then we ligated these two parts.
Construction of E. coli strain DH5α
Competent cells with luxR gene on pSB3K3 were constructed. Afterward, Plux-CmR on pSB6A1 was introduced into the cells.
The growth assay
In order to follow up the growth of the cells which Plux-CmR was introduced into, the change of O.D. (590nm) in both the presence and the absence of 3OC6HSL was measured.
(samples)
- Plux-CmR [http://partsregistry.org/Part:BBa_K395162 BBa_K395162]
- constitutive promoter (PlacIq)-CmR (positive control)[http://partsregistry.org/Part:BBa_K395165 BBa_K395165]
- promoterless-CmR (negative control)[http://partsregistry.org/Part:BBa_K395160 BBa_K395160]
LB medium was used as liquid culture. Antibiotics (ampicillin (Amp), kanamycin (Kan), and chloramphenicol (Cm)) were dissolved in distillation water and stored in 25 mg/ml concentration; their final concentration in the medium was adjusted to 50ug/ml, 30ug/ml, and 750ug/ml respectively.
- The seed cultures of the samples were inoculated from glycerol stock solution and grown separately overnight at 37°C in LB medium containing Amp & Kan.
- Then, we diluted the cultures to 100-fold by two types of 3 ml of fresh medium. One type was the medium containing 3OC6HSL (final concentration:100nM). Another medium contained DMSO as experiment control. Both of the cultures were incubated at 37°C
- 1 hour after dilution, each culture was separated into two new tubes. Next, 1.5 ml LB medium each containing Cm (final concentration:750ug/ml), Amp, Kan,3OC6HSL or DMSO was added to each tube.
- O.D.590 was measured every hour, up to 20 hours from dilution.
Reference
- Ying-jin Yuan et al PLoS 2010, e10619
- W. Shaw, et al Br. Med. Bull. 1984, 40, 36.
 "
"