Team:Tokyo Tech/Project/wolf coli/New Series of PompC
From 2010.igem.org
(→Overview of new OmpC promoter series) |
(→Result) |
||
| (79 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<div id="tf_menu"> | <div id="tf_menu"> | ||
| - | menu | + | <font size="5" color="#eb8300"><b><center>Project menu</center></b></font> |
| - | </ | + | |
| - | < | + | |
| - | = | + | <center> |
| - | + | <table id="table-01"> | |
| - | + | <tr> | |
| + | <td>[[Team:Tokyo_Tech|1 Graphic abstract]]<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <td>2 Apple reporter<br> | ||
| + | :[[Team:Tokyo_Tech/Project/Apple_Reporter|2-1 Color]] | ||
| + | :[[Team:Tokyo_Tech/Project/Apple_Reporter2|2-2 Fragrance]] | ||
| + | </td> | ||
| + | <tr> | ||
| + | <td>[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System|3 Artificial Cooperation System]]<br> | ||
| + | :[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System/lux_act_rep|3-1 lux activation/repression promoter]] | ||
| + | :[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System/Cm_assay|3-2 resistance gene activation device]] | ||
| + | :[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System/luxI_assay|3-3 ''lux''I Assay]] | ||
| + | :[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System/modeling|3-4 modeling]] | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>[[Team:Tokyo_Tech/Project/wolf_coli|4 Wolf coli overview]]<br> | ||
| + | :4-1 New seriesof P''ompC'' -YOU ARE HERE!- | ||
| + | :[[Team:Tokyo_Tech/Project/wolf_coli/lacIM1|4-2 lacIM1 for band-detect network ]] | ||
| + | :[[Team:Tokyo_Tech/Project/wolf_coli/System|4-3 Wolf coli system]] | ||
| + | </th> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| - | |||
| - | |||
| + | </div> <!-- tf_menu --> | ||
| + | <div id="tf_SubWrapper"> | ||
| + | <font size="5"><b>4-1 New series of P''ompC''</b></font> | ||
| + | ==Abstract== | ||
| + | In order to fine tune the Wolf coli system, we prepared a new series of ''OmpC'' promoter. The new series of promoters are P''ompC(C)'' [http://partsregistry.org/Part:BBa_K395301 BBa_K395301 ], P''ompC(CB)'' [http://partsregistry.org/Part:BBa_K395302 BBa_K395302 ]and P''ompC(CS1)'' [http://partsregistry.org/Part:BBa_K395303 BBa_K395303 ]. For measuring the strength of each promoter, we used GFP as a reporter. We have found that expression of GFP in ''OmpC(CB)'' and ''OmpC(CS1)'' promoters increased in high osmolarity medium. In contrast, under same conditions, there was no significant difference of GFP expression in ''OmpC(C)'' and ''OmpC(WT)'' promoters. | ||
| + | ==New BioBrick Parts== | ||
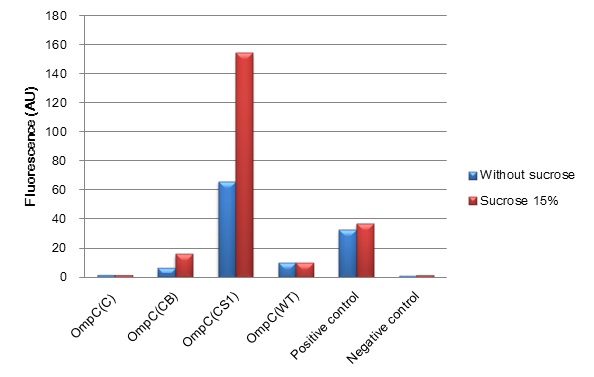
| + | [[Image:Tokyotech_ompc_graph.jpg|left|thumb|300px|Fig.4-1-1 The induction of new P''OmpC'' series in high osmolarity medium at 4 hours. This work is done by Thiprampai THAMAMONGOOD and Taichi NAKAMURA Tokyo Tech iGEM 2010]] | ||
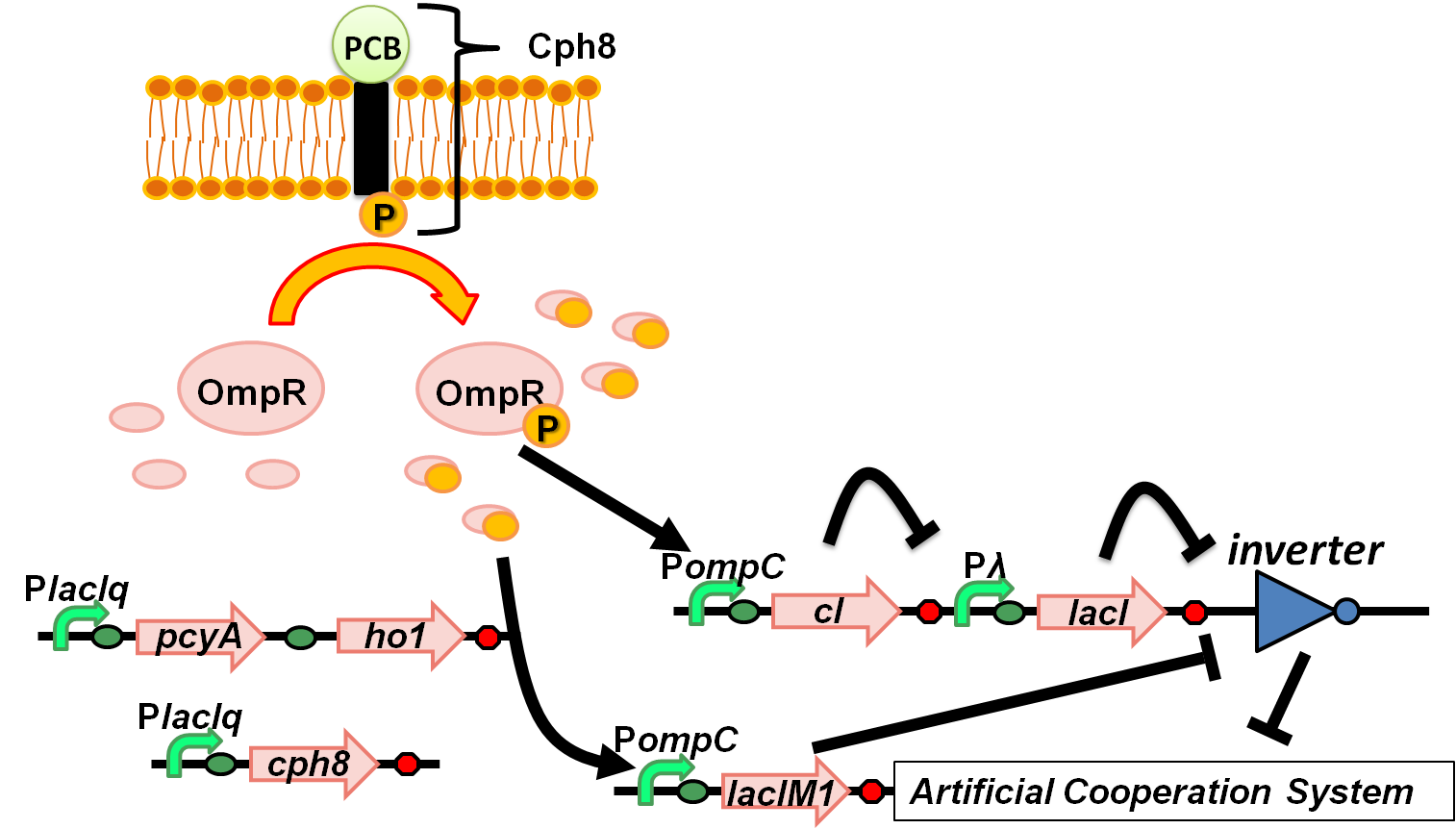
| + | [[Image:Tokyotech wolfcoli system_ver5.png|thumb|right|300px|Fig. 4-1-2. Overview of “Wolfcoli” system]] | ||
| Line 37: | Line 65: | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | (Fig. 4-1-1) We have succeeded in designing 2 new osmoregulative promoters, P''OmpC(CB)'' and P''OmpC(CS1)'', which can also be utilized in red-light-dependent gene expression network (Fig.4-1-2). | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ==Introduction== | ||
| + | *;What is ''OmpC'' promoter? | ||
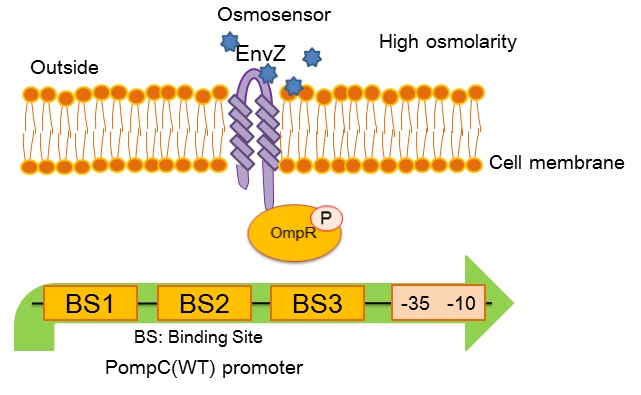
| + | [[image:Tokyotech_OmpC_wild_type.jpg|center|400px||thumb|Fig.4-1-3. PompC(WT) and the two-component system]] | ||
| + | ''OmpC(WT)'' promoter (BBa_R0082) is positively regulated by phosphorylated OmpR, a response regulator in EnvZ-OmpR osmosensing two-component system. The EnvZ-OmpR system of E. coli regulates the transcriptional activity of this promoter in response to change in extracellular osmolarity. The ''OmpC(WT)'' promoter consists of 3 distinct phosphorylated OmpR recognition sites which are binding site 1, binding site 2 and binding site 3. | ||
==Result== | ==Result== | ||
| - | ; | + | *;'''Construction of new series of ''OmpC'' promoters''' |
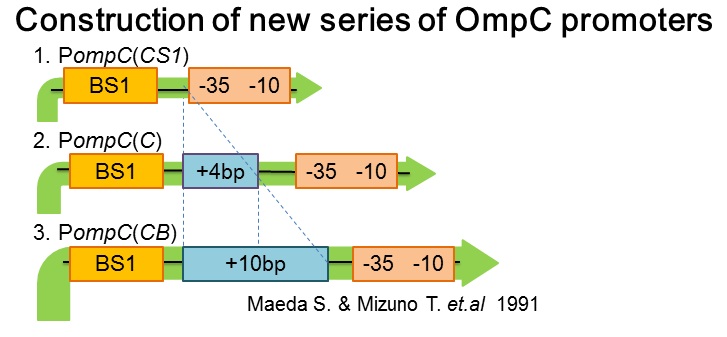
| - | [[ | + | [[image:Tokyotech_New_pompC_construction.jpg|center|400px|thumb|Fig.4-1-4. PompC(WT) and the two-component system]] |
| + | Three new promoters, P''ompC(CS1)'' BBa_395303, P''ompC(C)'' BBa_395301, P''ompC(CB)'' BBa_395302 were designed based on previous paper[1]. The second promoter, P''ompC(CS1)'', comprises only binding site1 fused to the canonical -35 and -10 region. P''ompC(C)'' is a composite of binding site 1 and additional 4 base pairs before the canonical -35 and -10 region. Likewise, P''ompC(CB)'' consists of binding site1 and the additional 10 base pairs before the -35 and -10 consensus sequence. | ||
| + | *; Characterization of new series of ''OmpC'' promoters | ||
| + | [[Image:Tokyotech_ompc_graph.jpg|thumb|center|400px|thumb|Fig.4-1-1 The induction of new'' OmpC'' series in high osmolarity medium at 4 hours. This work is done by Thiprampai THAMAMONGOOD and Taichi NAKAMURA ]] | ||
| + | After 4 hours of sucrose induction, transcriptional activity of P''ompC(CB)''-GFP and P''ompC(CS1)''-GFP increased 2.5-folds and 2.3-folds respectively. However, significant amount of leaky expression was found in P''ompC(CS1)''-GFP without induction. In contrast, under the same conditions, we found no significant difference of GFP expression in P''ompC(C)''-GFP. Although, P''ompC(WT)''-GFP also doesn't show difference in expression of GFP at 4 hours of induction, there was slightly higher GFP expression 1.7-folds occurred in the wild type at 2 hours. | ||
| + | ==Discussion== | ||
| + | The transcriptional activity of P''ompC(WT)''-GFP increased in particular period and declined after it reached the peak of expression. This result shows the relation to the study by the [http://partsregistry.org/Part:BBa_R0082:Experience Edinburgh iGEM 2009]. Moreover, we considereated that we can create varities of P''OmpC'' by combining OmpR binding sites on the upstream of the ''OmpC'' promoters with spacer sequence. | ||
| + | ==Conclusion== | ||
| + | According to the results, we have succeeded in comprising 2 new series of osmoregulative promoters. Furthermore, we expect that P''ompC(CB)'' would be a key component in responding to the middle light intensitys, “full moon light” and probably awaken our werewolf coli in the “full moon night” | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==Material and Method== | ==Material and Method== | ||
| - | + | ===Construction of E. coli strain MG1655=== | |
| - | Each new BioBrick part, P''ompC(C)''-GFP, P''ompC(CB)''-GFP, P''ompC(CS1)''-GFP and P''ompC(WT)''-GFP | + | Each new BioBrick part, P''ompC(C)''-GFP [http://partsregistry.org/Part:BBa_K395304 BBa_K395304 ], P''ompC(CB)''-GFP [http://partsregistry.org/Part:BBa_K395305 BBa_K395305 ], P''ompC(CS1)''-GFP [http://partsregistry.org/Part:BBa_K395306 BBa_K395306 ] and P''ompC(WT)''-GFP [http://partsregistry.org/Part:BBa_K395307 BBa_K395307 ] on pSB3K3 was introduced into E. coli strain MG1655. A strain containing plac<sup>q</sup>-GFP plasmid (BBa_J54202), a constitutive GFP expressive promoter, and promoterless GFP reporter plasmid (BBa_J54103) was used as a positive control and negative control respectively. |
| - | + | ===Medium=== | |
| - | Medium A, per liter, | + | Medium A, per liter, contains 7 g of nutrient broth, 1 g of yeast extract, 2 g of glycerol, 3.7 g of K2HPO4, and 1.3 g of KH2PO4 [3]. |
| - | + | ===Report assay=== | |
| - | In order to determine the strength of promoter parts in the reporter plasmid, the change of fluorescence intensities of the reporter stains in the presence and the absence of the inducer were measured. Overnight cultures of reporter strains grown at 37 °C in Medium A containing appropriated antibiotics were diluted at least 1:100 in the medium and incubated at 37 °C as fresh cultures. After their OD<sub>590</sub> reached 0.2, the fresh culture was diluted 1 : 3 into 2 ml of pre-warmed medium A. | + | In order to determine the strength of promoter parts in the reporter plasmid, the change of fluorescence intensities of the reporter stains in the presence and the absence of the inducer were measured. Overnight cultures of reporter strains grown at 37 °C in Medium A containing appropriated antibiotics were diluted at least 1:100 in the medium and incubated at 37 °C as fresh cultures. After their OD<sub>590</sub> reached 0.2, the fresh culture was diluted 1 : 3 into 2 ml of pre-warmed medium A. Under high osmolarity conditions, the cultures were diluted with sucrose supplemented medium to the final concentration of 15% (wt/vol)[2]. |
| - | + | ===Sample preparation for GFP measurement=== | |
After 4 hours of high ormolarity induction, 0.2 ml of each culture was moved to 2.0 ml eppendorf tube and then centrifuged for 1 min at 4°C, 9000 rpm. The supernatant was discarded from each tube by pipette. In order to adjust each sample to reach approximately same final OD<sub>590</sub>, appropriate amount of 1×PBS washing solution was added. The cell pellet in each tube was resuspended by vortexing. After all samples for GFP measurement were prepared, 150 μl of each sample was applied to 96-well plate and its fluorescence intensity was measured with fluorometer. The fluorescence intensity was calculated by dividing the measured raw fluorescence intensity by the adjusted OD<sub>590</sub>. | After 4 hours of high ormolarity induction, 0.2 ml of each culture was moved to 2.0 ml eppendorf tube and then centrifuged for 1 min at 4°C, 9000 rpm. The supernatant was discarded from each tube by pipette. In order to adjust each sample to reach approximately same final OD<sub>590</sub>, appropriate amount of 1×PBS washing solution was added. The cell pellet in each tube was resuspended by vortexing. After all samples for GFP measurement were prepared, 150 μl of each sample was applied to 96-well plate and its fluorescence intensity was measured with fluorometer. The fluorescence intensity was calculated by dividing the measured raw fluorescence intensity by the adjusted OD<sub>590</sub>. | ||
| + | |||
==References== | ==References== | ||
1. Maeda S. & Mizuno T. Activation of the Osmoregulated ''ompC'' Gene by the ''OmpR'' Protein in Escherichia coli. J.Biochem 1991;110,324-27 <br> | 1. Maeda S. & Mizuno T. Activation of the Osmoregulated ''ompC'' Gene by the ''OmpR'' Protein in Escherichia coli. J.Biochem 1991;110,324-27 <br> | ||
Latest revision as of 03:34, 28 October 2010
4-1 New series of PompC
Contents |
Abstract
In order to fine tune the Wolf coli system, we prepared a new series of OmpC promoter. The new series of promoters are PompC(C) [http://partsregistry.org/Part:BBa_K395301 BBa_K395301 ], PompC(CB) [http://partsregistry.org/Part:BBa_K395302 BBa_K395302 ]and PompC(CS1) [http://partsregistry.org/Part:BBa_K395303 BBa_K395303 ]. For measuring the strength of each promoter, we used GFP as a reporter. We have found that expression of GFP in OmpC(CB) and OmpC(CS1) promoters increased in high osmolarity medium. In contrast, under same conditions, there was no significant difference of GFP expression in OmpC(C) and OmpC(WT) promoters.
New BioBrick Parts
(Fig. 4-1-1) We have succeeded in designing 2 new osmoregulative promoters, POmpC(CB) and POmpC(CS1), which can also be utilized in red-light-dependent gene expression network (Fig.4-1-2).
Introduction
- What is OmpC promoter?
OmpC(WT) promoter (BBa_R0082) is positively regulated by phosphorylated OmpR, a response regulator in EnvZ-OmpR osmosensing two-component system. The EnvZ-OmpR system of E. coli regulates the transcriptional activity of this promoter in response to change in extracellular osmolarity. The OmpC(WT) promoter consists of 3 distinct phosphorylated OmpR recognition sites which are binding site 1, binding site 2 and binding site 3.
Result
- Construction of new series of OmpC promoters
Three new promoters, PompC(CS1) BBa_395303, PompC(C) BBa_395301, PompC(CB) BBa_395302 were designed based on previous paper[1]. The second promoter, PompC(CS1), comprises only binding site1 fused to the canonical -35 and -10 region. PompC(C) is a composite of binding site 1 and additional 4 base pairs before the canonical -35 and -10 region. Likewise, PompC(CB) consists of binding site1 and the additional 10 base pairs before the -35 and -10 consensus sequence.
- Characterization of new series of OmpC promoters
After 4 hours of sucrose induction, transcriptional activity of PompC(CB)-GFP and PompC(CS1)-GFP increased 2.5-folds and 2.3-folds respectively. However, significant amount of leaky expression was found in PompC(CS1)-GFP without induction. In contrast, under the same conditions, we found no significant difference of GFP expression in PompC(C)-GFP. Although, PompC(WT)-GFP also doesn't show difference in expression of GFP at 4 hours of induction, there was slightly higher GFP expression 1.7-folds occurred in the wild type at 2 hours.
Discussion
The transcriptional activity of PompC(WT)-GFP increased in particular period and declined after it reached the peak of expression. This result shows the relation to the study by the [http://partsregistry.org/Part:BBa_R0082:Experience Edinburgh iGEM 2009]. Moreover, we considereated that we can create varities of POmpC by combining OmpR binding sites on the upstream of the OmpC promoters with spacer sequence.
Conclusion
According to the results, we have succeeded in comprising 2 new series of osmoregulative promoters. Furthermore, we expect that PompC(CB) would be a key component in responding to the middle light intensitys, “full moon light” and probably awaken our werewolf coli in the “full moon night”
Material and Method
Construction of E. coli strain MG1655
Each new BioBrick part, PompC(C)-GFP [http://partsregistry.org/Part:BBa_K395304 BBa_K395304 ], PompC(CB)-GFP [http://partsregistry.org/Part:BBa_K395305 BBa_K395305 ], PompC(CS1)-GFP [http://partsregistry.org/Part:BBa_K395306 BBa_K395306 ] and PompC(WT)-GFP [http://partsregistry.org/Part:BBa_K395307 BBa_K395307 ] on pSB3K3 was introduced into E. coli strain MG1655. A strain containing placq-GFP plasmid (BBa_J54202), a constitutive GFP expressive promoter, and promoterless GFP reporter plasmid (BBa_J54103) was used as a positive control and negative control respectively.
Medium
Medium A, per liter, contains 7 g of nutrient broth, 1 g of yeast extract, 2 g of glycerol, 3.7 g of K2HPO4, and 1.3 g of KH2PO4 [3].
Report assay
In order to determine the strength of promoter parts in the reporter plasmid, the change of fluorescence intensities of the reporter stains in the presence and the absence of the inducer were measured. Overnight cultures of reporter strains grown at 37 °C in Medium A containing appropriated antibiotics were diluted at least 1:100 in the medium and incubated at 37 °C as fresh cultures. After their OD590 reached 0.2, the fresh culture was diluted 1 : 3 into 2 ml of pre-warmed medium A. Under high osmolarity conditions, the cultures were diluted with sucrose supplemented medium to the final concentration of 15% (wt/vol)[2].
Sample preparation for GFP measurement
After 4 hours of high ormolarity induction, 0.2 ml of each culture was moved to 2.0 ml eppendorf tube and then centrifuged for 1 min at 4°C, 9000 rpm. The supernatant was discarded from each tube by pipette. In order to adjust each sample to reach approximately same final OD590, appropriate amount of 1×PBS washing solution was added. The cell pellet in each tube was resuspended by vortexing. After all samples for GFP measurement were prepared, 150 μl of each sample was applied to 96-well plate and its fluorescence intensity was measured with fluorometer. The fluorescence intensity was calculated by dividing the measured raw fluorescence intensity by the adjusted OD590.
References
1. Maeda S. & Mizuno T. Activation of the Osmoregulated ompC Gene by the OmpR Protein in Escherichia coli. J.Biochem 1991;110,324-27
2. Batchelor E. & Goulian M. Imaging OmpR localization in Escherichia coli. Molecular Microbiology. 2006;59(6),1767-78
3. Kawaji H. Influence of Molecular Size and Osmolarity of Sugars and Dextrans on the Synthesis of Outer Membrane Proteins 0-8 and 0-9 of Escherichia coli K-12. J.Bacteriology 1979;140(3), 843-47
 "
"