Team:Tokyo Tech/Project/Apple Reporter
From 2010.igem.org
(→zeaxanthin synthesis) |
(→zeaxanthin synthesis) |
||
| (83 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
<table id="table-01"> | <table id="table-01"> | ||
<tr> | <tr> | ||
| - | <td>[[Team:Tokyo_Tech|1 | + | <td>[[Team:Tokyo_Tech|1 Graphic abstract]]<br> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 26: | Line 26: | ||
<tr> | <tr> | ||
<td>[[Team:Tokyo_Tech/Project/wolf_coli|4 wolfcoli overview]]<br> | <td>[[Team:Tokyo_Tech/Project/wolf_coli|4 wolfcoli overview]]<br> | ||
| - | :[[Team:Tokyo_Tech/Project/wolf_coli/New_Series_of_PompC|4-1 | + | :[[Team:Tokyo_Tech/Project/wolf_coli/New_Series_of_PompC|4-1 New series of P''ompC'']] |
:[[Team:Tokyo_Tech/Project/wolf_coli/lacIM1|4-2 lacIM1 for band-detect network ]] | :[[Team:Tokyo_Tech/Project/wolf_coli/lacIM1|4-2 lacIM1 for band-detect network ]] | ||
| - | :[[Team:Tokyo_Tech/Project/wolf_coli/System|4-3 | + | :[[Team:Tokyo_Tech/Project/wolf_coli/System|4-3 Wolf coli system]] |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 42: | Line 42: | ||
<!-- ここから始まるよ--> | <!-- ここから始まるよ--> | ||
==Abstract== | ==Abstract== | ||
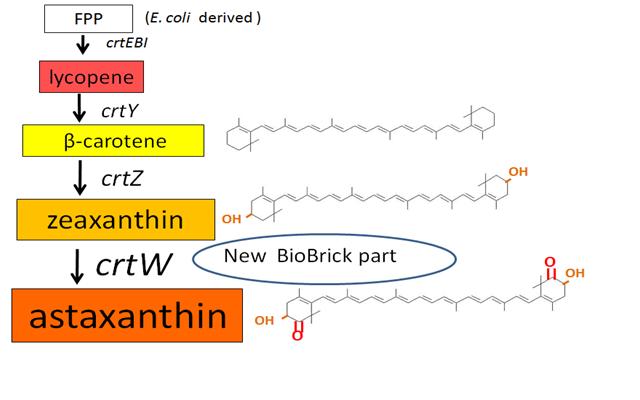
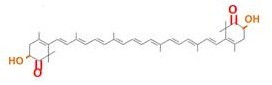
| - | Our team synthesized β-carotene, zeaxanthin and astaxanthin under several conditions (strain, | + | Our team synthesized β-carotene, zeaxanthin and astaxanthin under several conditions (strain, plasmid copy number,…etc) to confirm the functions of BioBrick parts designed by previous iGEM teams. Particularly, '''astaxanthin is the final metabolite of the carotenoid synthetic pathway.''' |
| - | We synthesized β-carotene and zeaxanthin by assembling ''crtEBIY'' and ''crtZ'' of existing BioBrick parts. '''Though the part ''crtZ'' has been unconfirmed, we confirmed it for the first time in iGEM.''' | + | We synthesized β-carotene and zeaxanthin by assembling ''crtEBIY'' and ''crtZ'' of existing BioBrick parts. '''Though the part ''crtZ'' has been unconfirmed to produce zeaxanthin, we confirmed it for the first time in iGEM.''' |
In addition, we succeeded in synthesizing astaxanthin by introducing '''the new BioBrick part ''crtW'''''. Therefore, the carotenoid synthetic pathway reached the end. | In addition, we succeeded in synthesizing astaxanthin by introducing '''the new BioBrick part ''crtW'''''. Therefore, the carotenoid synthetic pathway reached the end. | ||
| - | |||
| + | [[Image:tokyotech_Fig. 1.1.1. carotenoid synthetic pathway.jpg|390px|left|thumb|Fig. 2-1-1. carotenoid synthetic pathway]] | ||
| - | + | [[Image:TLC work3.jpeg|right|210px|thumb|Fig.2-1-2.TLC worked by Yumiko Kinoshita]] | |
| - | + | ||
| - | [[Image: | + | |
| - | + | ||
| - | + | ||
[https://2010.igem.org/Team:Tokyo_Tech/Parts →our BioBrick parts] | [https://2010.igem.org/Team:Tokyo_Tech/Parts →our BioBrick parts] | ||
| + | [http://partsregistry.org/Part:BBa_K395704 →BBa_395704] | ||
| + | [http://partsregistry.org/Part:BBa_K395706 →BBa_395706] | ||
==Introduction== | ==Introduction== | ||
| - | Carotenoids are natural organic pigments in plants and bacteria. Synthetic carotenoid pigments colored yellow, red or orange represent about 15-25% of the cost of production of commercial feed. Carotenoids are terpenoid based on a structure having the chemical formula C<sub>40</sub>H<sub>56</sub>. There are about 600 carotenoids, which perform a wide range of functions. (For example | + | Carotenoids are natural organic pigments in plants and bacteria. Synthetic carotenoid pigments colored yellow, red or orange represent about 15-25% of the cost of production of commercial feed. Carotenoids are terpenoid based on a structure having the chemical formula C<sub>40</sub>H<sub>56</sub>. There are about 600 carotenoids, which perform a wide range of functions. (For example, acting as antioxidants[https://2010.igem.org/Team:Tokyo_Tech/Project/Apple_Reporter#Reference], and as precursors to other organic compounds.) '''Among carotenoids, astaxanthin is not known to be converted to Vitamin A, which is converted from most of carotenoids and is toxic when its concentration too high.''' |
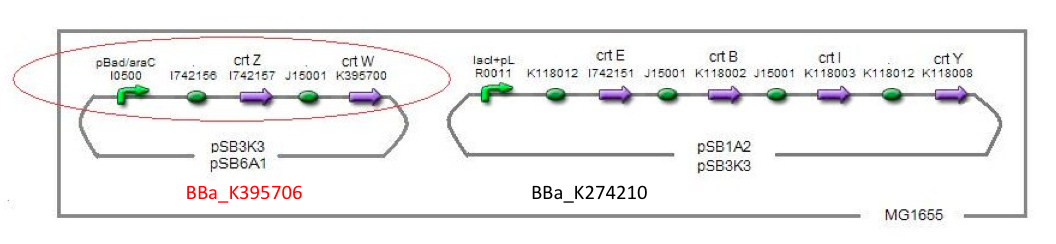
Previous year, our team has completed melanin synthetic pathway. Since the carotenoid synthetic pathway is huge, BioBrick Registry doesn’t cover all parts to complete the pathway. '''This year, we introduced a new BioBrick part to synthesize more various kinds of carotenoids. And we succeeded in synthesizing astaxanthin, one of the final metabolite of the pathway.''' | Previous year, our team has completed melanin synthetic pathway. Since the carotenoid synthetic pathway is huge, BioBrick Registry doesn’t cover all parts to complete the pathway. '''This year, we introduced a new BioBrick part to synthesize more various kinds of carotenoids. And we succeeded in synthesizing astaxanthin, one of the final metabolite of the pathway.''' | ||
| - | + | [[Image:tokyotech_tokyotech_Fig. 1.1.5. note.jpg|500px|center|thumb|Fig. 2-1-3. note]] | |
| - | + | ||
| - | + | ||
| - | [[Image:tokyotech_tokyotech_Fig. 1.1.5. note.jpg|500px|center| | + | |
| - | + | ||
==Results== | ==Results== | ||
===β-carotene synthesis=== | ===β-carotene synthesis=== | ||
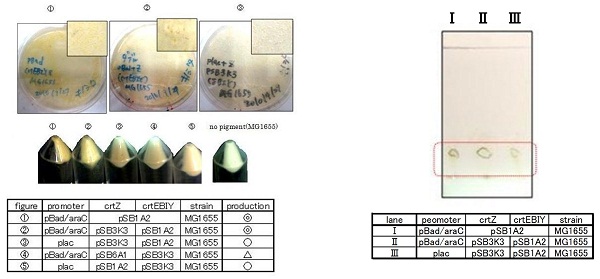
| - | [[Image:tokyotech_Fig. 1.1.2.1. β-carotene.jpg|right|200px|Fig. 1 | + | [[Image:tokyotech_Fig. 1.1.2.1. β-carotene.jpg|right|200px|thumb|Fig.2-1-4.β-carotene]] |
| - | Our team synthesized β-carotene under several conditions to confirm the functions of BioBrick parts designed by Cambridge 2009. | + | Our team synthesized β-carotene under several conditions to confirm the functions of BioBrick parts designed by Cambridge 2009. (we call it "crtEBIY plasmids".) ([https://2010.igem.org/Team:Tokyo_Tech/Project/Apple_Reporter#appendix See more...]) |
===zeaxanthin synthesis=== | ===zeaxanthin synthesis=== | ||
| - | [[Image:tokyotech_Fig. 1.1.6. zeaxanthin.jpg|right|200px|Fig. 1. | + | [[Image:tokyotech_Fig. 1.1.6. zeaxanthin.jpg|right|200px|thumb|Fig.2-1-5zeaxanthin]] |
| + | Our team tried to synthesize zeaxanthin by using the constructed ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K395704 BBa_K395704])part to confirm the activity of BioBrick parts designed by Cambredge 2009 and Edinburgh 2007. We also confirmed zeaxanthin by TLC (Fig.2-1-6 spot III). | ||
| - | |||
| - | |||
| - | + | By introduction of ''crtZ'' gene to the ''crtEBIY'' plasmid (we call this construct “single plasmid construct”, "''crtZ-crtEBIY''".), new spot appered on TLC plate (Fig.2-1-6 spot III). The spot shows lower Rf value than that derived from ''crtEBIY'' (Fig.2-1-6 spot II). The Rf values of spots derived from ''crtEBIY'' and ''crtZ-crtEBIY'' corresponded to reported values of β-caroten and Zeaxanthin, respectively.[https://2010.igem.org/Team:Tokyo_Tech/Project/Apple_Reporter#Reference] Thus we could characterize zeaxanthin production activity of crtZ. In addition, caracterization of the part ''crtZ'' is the first work in iGEM. | |
| - | + | ||
| - | + | ||
| - | The | + | |
| - | |||
| - | + | In order to maximize zeaxanthin production, we designed some combinations of construct. We thought of another type of zeaxanthin synthetic construct where ''“crtZ”'' (BBa_I742158) and ''“crtEBIY”'' (BBa_K274210) were separated on different plasmids. (we call this type of constructs “double plasmids construct.”) | |
| - | |||
| - | |||
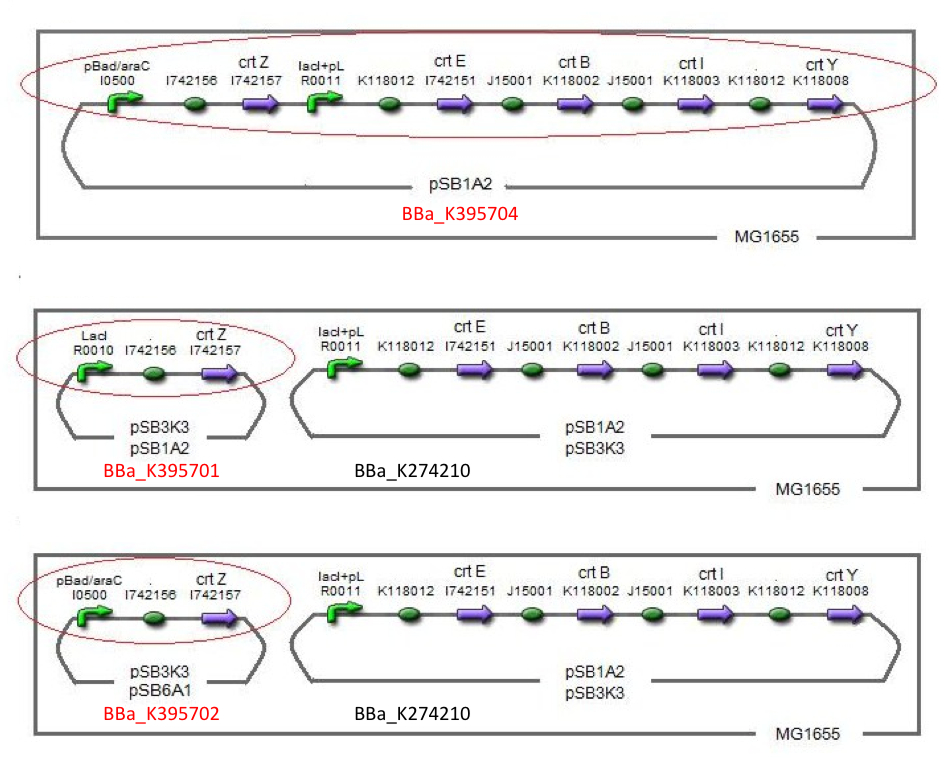
| + | After all, We made five combinations shown in table<1>, including [http://partsregistry.org/wiki/index.php?title=Part:BBa_K395701 BBa_K395701], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K395702 BBa_K395702] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K395704 BBa_K395704]. | ||
| + | Constructs containing pBad/araC promoter on high-copy vector show the best production and double plasmids construct shows as much production as single plasmid construct under same promoter combination. Construct with pBad/araC promoter shows better production than that with plac promoter. Construct that ''crtEBIY'' on high-copy vector and ''crtZ'' on low-copy vector shows as much expression as construct that ''crtEBIY'' on low-copy vector and ''crtZ'' on high-copy vector.(Actually, 3 showed a little more production than 5.) | ||
| - | |||
| - | + | For TLC, We selected the best three samples including the sample derived from single plasmid construct . | |
| - | + | In the consequence of TLC, we could confirm that these three samples were zeaxanthin. | |
| - | |||
| - | |||
| - | |||
| - | + | [[Image:S1-zeaxanthin.jpg|left|300px|thumb|S1 : zeaxanthin expression components]] | |
| - | + | ||
| - | + | [[Image:TLC work3.jpeg|right|300px|thumb|Fig.2-1-6 zeaxanthin TLC worked by Yumiko Kinoshita]] | |
| + | [[Image:Tokyotech_zeaxanthin_synthesis.jpg|center|500px|thumb|Fig.2-1-7 table<1> zeaxanthin worked by Yumiko Kinoshita]] | ||
| - | [[Image:tokyotech_Fig. | + | ===astaxanthin synthesis=== |
| + | [[Image:tokyotech_Fig. 1.1.9. zeaxanthin.jpg|right|200px|thumb|Fig.2-1-8 astaxanthin]] | ||
| + | Our team synthesized astaxanthin for the first time in iGEM. Thus the carotenoid synthetic pathway reached the end. This work was done by assembling the new BioBrick part ''crtW'' and zeaxanthin synthesis constructs we designed. From the result of “zeaxanthin synthesis”, we decided to use pBad/araC promoter and double plasmids construct. | ||
| + | After constructing the new BioBrick device ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K395706 BBa_K395706]) to synthesize astaxanthin, we characterize the function under the condition I in table<2>. Thus we confirmed astaxanthin by TLC (Fig2-1-6 spot IV). | ||
| - | [ | + | By introduction of ''crtW'' gene to the ''crtZ'' plasmid (the second construct of five combinations in “zeaxanthin synthesis”,having pBad/araC promoter), new spot ((Fig2-1-6 spot IV) appeared on TLC plate. The spot shows lower Rf value than that derived from ''crtEBIY'' (II) and a little higher Rf value than that derived from ''crtZ-crtW''(III). All the Rf values of spots corresponded to reported values respectively.[https://2010.igem.org/Team:Tokyo_Tech/Project/Apple_Reporter#Reference [2]] Thus we succeeded in characterizing astaxanthin production activity of ''crtW''. ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K395705 BBa_K395705], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K395706 BBa_K395706]) |
| - | |||
| - | + | For the same reason as “zeaxanthin synthesis”, we examined under several conditions →table<2>. | |
| - | + | In the consequence of TLC, we could confirm that these four samples were astaxanthin. | |
| - | + | ||
| + | Not only astaxanthin but also another spots were produced at II. | ||
| + | We regarded them as intermediary metabolites between zeaxanthin and astaxanthin. | ||
| + | The activity of pBad/araC promoter is measured in detail on another page. [https://2010.igem.org/Team:Tokyo_Tech/Project/wolf_coli/lacIM1 →click] | ||
| + | [[Image:S2-astaxanthin.jpg|left|300px|thumb|S2 : astaxanthin expression component]] | ||
| + | [[Image:TLC work3.jpeg|right|300px|thumb|Fig.2-1-6 TLC astaxanthin worked by Yumiko Kinoshita]] | ||
| + | [[Image:Tokyotech_astaxanthin_synthesis.jpg|center|500px|thumb|Fig.2-1-9 astaxanthin worked by Yumiko Kinoshita]] | ||
| - | |||
| - | + | acetone extract and vacuum concentration [https://2010.igem.org/Team:Tokyo_Tech/Project/Apple_Reporter#Materials_and_Methods →protocol] | |
==Reference== | ==Reference== | ||
| - | [1]N.M. Sachindra, et al. Carotenoids in crabs from marine and fresh waters of India. LWT 38 (2005) 221–225 | + | [1] Yousry M. Antioxidant Activities of Astaxanthin and Related Carotenoids. J. Agric. Food Chem. 2000, 48, 1150-1154 |
| + | [2]N.M. Sachindra, et al. Carotenoids in crabs from marine and fresh waters of India. LWT 38 (2005) 221–225 | ||
| - | [ | + | [3]MISAWA N, et al. Structure and Functional Analysis of a Marine Bacterial Carotenoid Biosynthesis Gene Cluster and Astaxanthin Biosynthetic Pathway Proposed at the Gene Level. JOURNAL OF BACTERIOLOGY, Nov. 1995, p. 6575–6584 Vol. 177, No. 22 |
| - | [ | + | [4]Seon-Kang C, et al. Characterization of bacterial β-carotene Ketolases, CrtW, |
| + | from Mrine Bacteria by Complementation Analysis in ''Escherichia coli''. MARINE BIOTECHNOLOGY(2005) 7, 515-522 | ||
| - | [ | + | [5]Seon-Kang C, et al. Characterization of bacterial β-carotene 3,3′-hydroxylases, CrtZ, |
and P450 in astaxanthin biosynthetic pathway and adonirubin | and P450 in astaxanthin biosynthetic pathway and adonirubin | ||
production by gene combination in ''Escherichia coli''. Appl Microbiol Biotechnol (2006) 72 | production by gene combination in ''Escherichia coli''. Appl Microbiol Biotechnol (2006) 72 | ||
| - | [ | + | |
| + | [6]Nishizaki T, et al. Metabolic engineering of carotenoid biosynthesis in ''Escherichia coli'' by ordered gene assembly in Bacillus subtilis. Appl Environ Microbiol. 2007 Feb | ||
| + | |||
| + | [7]Arunkumar K, et al. Structure of a carotenoid gene cluster from Pantoea sp. strain C1B1Y and characterization of β-carotene hydroxylase(crtZ)gene by functional complementation in ''Escherichia coli''. Carotenoid Science Vol.11 (2007) | ||
==appendix== | ==appendix== | ||
===β-carotene synthesis=== | ===β-carotene synthesis=== | ||
| - | [[Image:tokyotech_Fig. 1.1.2.1. β-carotene.jpg|right|200px|Fig. 1 | + | [[Image:tokyotech_Fig. 1.1.2.1. β-carotene.jpg|right|200px|thumb|Fig.2-1-4 β-carotene]] |
Our team synthesized β-carotene under several conditions to confirm the activity of BioBrick parts designed by Cambredge 2009. | Our team synthesized β-carotene under several conditions to confirm the activity of BioBrick parts designed by Cambredge 2009. | ||
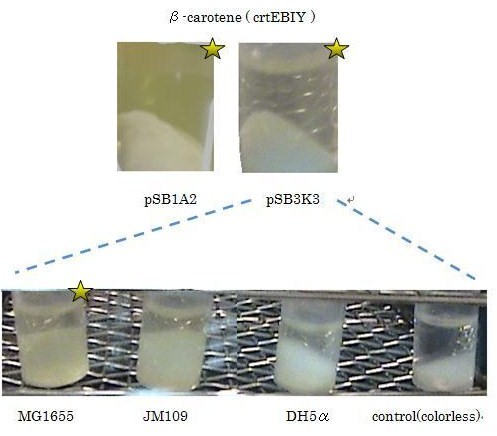
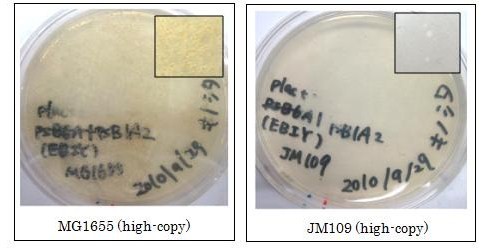
| - | We used ''crtEBIY'' (BBa_K274210) to synthesize β-carotene. We compared the plasmids containing pSB3K3 (low-copy vector) with the plasmids containing pSB1A2 (high-copy vector) from the production. We also introduced the low-copy plasmid into ''E. Coli'' strain MG1655, JM109 and DH5& | + | We used ''crtEBIY'' (BBa_K274210) to synthesize β-carotene. We compared the plasmids containing pSB3K3 (low-copy vector) with the plasmids containing pSB1A2 (high-copy vector) from the production. We also introduced the low-copy plasmid into ''E. Coli'' strain MG1655, JM109 and DH5α respectively. |
We found that MG1655 and JM109 produce more β-carotene than DH5α in liquid culture and that MG1655 grow better than JM109 on plate culture. These results indicated that MG1655 is the best of the three strains to treat. For this reason, we decided to use only MG1655 to extract pigment. | We found that MG1655 and JM109 produce more β-carotene than DH5α in liquid culture and that MG1655 grow better than JM109 on plate culture. These results indicated that MG1655 is the best of the three strains to treat. For this reason, we decided to use only MG1655 to extract pigment. | ||
| Line 175: | Line 171: | ||
[https://2010.igem.org/Team:Tokyo_Tech/Project/Apple_Reporter#Materials_and_Methods →protocol] | [https://2010.igem.org/Team:Tokyo_Tech/Project/Apple_Reporter#Materials_and_Methods →protocol] | ||
| - | [[Image:tokyotech_Fig. 1.1.3. β-carotene extract.jpg|300px|left|thumb|Fig. 2 | + | [[Image:tokyotech_Fig. 1.1.3. β-carotene extract.jpg|300px|left|thumb|Fig.2-1-10. β-carotene extract /worked by Yumiko Kinoshita]] |
| - | [[Image:tokyotech_Fig. 1.1.4. β-carotene plate.jpg|300px|right|thumb|Fig. 2- | + | [[Image:tokyotech_Fig. 1.1.4. β-carotene plate.jpg|300px|right|thumb|Fig.2-1-11. β-carotene plate /worked by Yumiko Kinoshita]] |
| + | |||
| + | |||
| + | [[Image:Βカロテン2.png|200px|center|thumb|Fig.2-1-12. β-carotene table /worked by Yumiko Kinoshita]] | ||
==Materials and Methods== | ==Materials and Methods== | ||
| - | ===The work flow to TLC from culture=== | + | ===The work flow to TLC from culture[https://2010.igem.org/Team:Tokyo_Tech/Project/Apple_Reporter#Reference],[https://2010.igem.org/Team:Tokyo_Tech/Project/Apple_Reporter#Reference]=== |
| + | ====Material==== | ||
| + | ''E. coli'' : strain MG1655 | ||
| + | |||
| + | reagent : acetone, methanol, chloroform, hexane | ||
| + | |||
| + | TLC plate : silicagel 60F<sub>254</sub> | ||
| + | |||
| + | ====culture==== | ||
1. The cells were cultured over night. | 1. The cells were cultured over night. | ||
| - | 2. 50ul bacterial | + | 2. 50ul bacterial solution was taken from culture prepared in 1 and moved into 50ml LB culture with appropriate antibiotic. Inducer was added, if necessary. |
| - | Inducer was added,if necessary. | + | |
| - | 3. The cells were incubated at | + | 3. The cells were incubated at 37°C, 24hour. |
| + | ====gathering ''E. coli'' and extract pigment==== | ||
4. After the O.D reach 2.0, culture was centrifuged for 10minutes at | 4. After the O.D reach 2.0, culture was centrifuged for 10minutes at | ||
| - | + | 4°C, 6000rpm,10min | |
| - | + | ||
5. Supernatant was removed by decanting. The remainder was added with 1 ml ddH<sub>2</sub>0 and vortexed. | 5. Supernatant was removed by decanting. The remainder was added with 1 ml ddH<sub>2</sub>0 and vortexed. | ||
6. Moved into 2ml tube by using P1000 pipetter at least twice. | 6. Moved into 2ml tube by using P1000 pipetter at least twice. | ||
| - | 7. Solution is centrifuged for 20minutes at | + | 7. Solution is centrifuged for 20minutes at 4°C, 14000rpm |
8. Supernatant was removed completely using pipetter. About 100ul of pellet is obtained. | 8. Supernatant was removed completely using pipetter. About 100ul of pellet is obtained. | ||
| Line 202: | Line 209: | ||
9. Pellet is added with 500ul acetone, and vortexed for 10 minutes to extract the pigment. | 9. Pellet is added with 500ul acetone, and vortexed for 10 minutes to extract the pigment. | ||
| - | 10. Solution is centrifuged for 20minutes at | + | 10. Solution is centrifuged for 20minutes at 4°C, 14000rpm |
11. Acetone solution was replaced into 2 ml tube. | 11. Acetone solution was replaced into 2 ml tube. | ||
| + | ====vacuum and concentrate==== | ||
12. Acetone was removed using rotary evaporator. | 12. Acetone was removed using rotary evaporator. | ||
| - | 13. 50 ul of Chloroform/Methanol | + | 13. 50 ul of Chloroform/Methanol(weight ratio of 9:1) solution was added. |
14. Sample was vortexed and spinned down. | 14. Sample was vortexed and spinned down. | ||
| Line 214: | Line 222: | ||
15. The water layer was removed. | 15. The water layer was removed. | ||
| - | 16. Spot the sample onto TLC silica gel plate, and developed by acetone/hexane | + | ====TLC==== |
| + | 16. Spot the sample onto TLC silica gel plate, and developed by acetone/hexane(weight ratio of 1:3) solution. | ||
17. Spot was observed under visible light. | 17. Spot was observed under visible light. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Latest revision as of 03:57, 28 October 2010
2-1 Color
Contents |
Abstract
Our team synthesized β-carotene, zeaxanthin and astaxanthin under several conditions (strain, plasmid copy number,…etc) to confirm the functions of BioBrick parts designed by previous iGEM teams. Particularly, astaxanthin is the final metabolite of the carotenoid synthetic pathway.
We synthesized β-carotene and zeaxanthin by assembling crtEBIY and crtZ of existing BioBrick parts. Though the part crtZ has been unconfirmed to produce zeaxanthin, we confirmed it for the first time in iGEM.
In addition, we succeeded in synthesizing astaxanthin by introducing the new BioBrick part crtW. Therefore, the carotenoid synthetic pathway reached the end.
→our BioBrick parts [http://partsregistry.org/Part:BBa_K395704 →BBa_395704] [http://partsregistry.org/Part:BBa_K395706 →BBa_395706]
Introduction
Carotenoids are natural organic pigments in plants and bacteria. Synthetic carotenoid pigments colored yellow, red or orange represent about 15-25% of the cost of production of commercial feed. Carotenoids are terpenoid based on a structure having the chemical formula C40H56. There are about 600 carotenoids, which perform a wide range of functions. (For example, acting as antioxidants[1], and as precursors to other organic compounds.) Among carotenoids, astaxanthin is not known to be converted to Vitamin A, which is converted from most of carotenoids and is toxic when its concentration too high.
Previous year, our team has completed melanin synthetic pathway. Since the carotenoid synthetic pathway is huge, BioBrick Registry doesn’t cover all parts to complete the pathway. This year, we introduced a new BioBrick part to synthesize more various kinds of carotenoids. And we succeeded in synthesizing astaxanthin, one of the final metabolite of the pathway.
Results
β-carotene synthesis
Our team synthesized β-carotene under several conditions to confirm the functions of BioBrick parts designed by Cambridge 2009. (we call it "crtEBIY plasmids".) (See more...)
zeaxanthin synthesis
Our team tried to synthesize zeaxanthin by using the constructed ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K395704 BBa_K395704])part to confirm the activity of BioBrick parts designed by Cambredge 2009 and Edinburgh 2007. We also confirmed zeaxanthin by TLC (Fig.2-1-6 spot III).
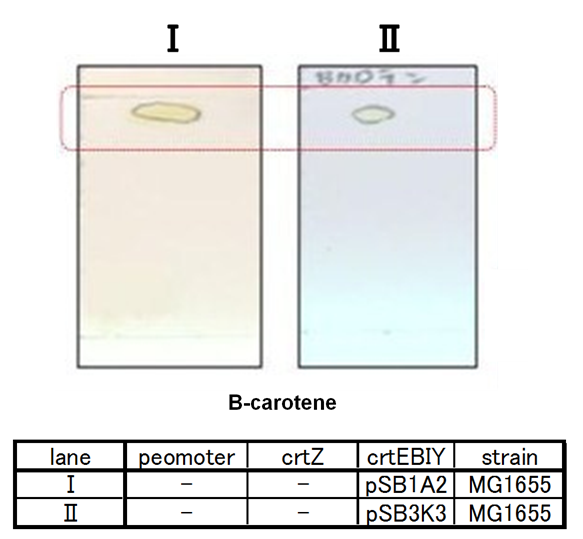
By introduction of crtZ gene to the crtEBIY plasmid (we call this construct “single plasmid construct”, "crtZ-crtEBIY".), new spot appered on TLC plate (Fig.2-1-6 spot III). The spot shows lower Rf value than that derived from crtEBIY (Fig.2-1-6 spot II). The Rf values of spots derived from crtEBIY and crtZ-crtEBIY corresponded to reported values of β-caroten and Zeaxanthin, respectively.[2] Thus we could characterize zeaxanthin production activity of crtZ. In addition, caracterization of the part crtZ is the first work in iGEM.
In order to maximize zeaxanthin production, we designed some combinations of construct. We thought of another type of zeaxanthin synthetic construct where “crtZ” (BBa_I742158) and “crtEBIY” (BBa_K274210) were separated on different plasmids. (we call this type of constructs “double plasmids construct.”)
After all, We made five combinations shown in table<1>, including [http://partsregistry.org/wiki/index.php?title=Part:BBa_K395701 BBa_K395701], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K395702 BBa_K395702] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K395704 BBa_K395704].
Constructs containing pBad/araC promoter on high-copy vector show the best production and double plasmids construct shows as much production as single plasmid construct under same promoter combination. Construct with pBad/araC promoter shows better production than that with plac promoter. Construct that crtEBIY on high-copy vector and crtZ on low-copy vector shows as much expression as construct that crtEBIY on low-copy vector and crtZ on high-copy vector.(Actually, 3 showed a little more production than 5.)
For TLC, We selected the best three samples including the sample derived from single plasmid construct .
In the consequence of TLC, we could confirm that these three samples were zeaxanthin.
astaxanthin synthesis
Our team synthesized astaxanthin for the first time in iGEM. Thus the carotenoid synthetic pathway reached the end. This work was done by assembling the new BioBrick part crtW and zeaxanthin synthesis constructs we designed. From the result of “zeaxanthin synthesis”, we decided to use pBad/araC promoter and double plasmids construct. After constructing the new BioBrick device ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K395706 BBa_K395706]) to synthesize astaxanthin, we characterize the function under the condition I in table<2>. Thus we confirmed astaxanthin by TLC (Fig2-1-6 spot IV).
By introduction of crtW gene to the crtZ plasmid (the second construct of five combinations in “zeaxanthin synthesis”,having pBad/araC promoter), new spot ((Fig2-1-6 spot IV) appeared on TLC plate. The spot shows lower Rf value than that derived from crtEBIY (II) and a little higher Rf value than that derived from crtZ-crtW(III). All the Rf values of spots corresponded to reported values respectively.[2] Thus we succeeded in characterizing astaxanthin production activity of crtW. ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K395705 BBa_K395705], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K395706 BBa_K395706])
For the same reason as “zeaxanthin synthesis”, we examined under several conditions →table<2>.
In the consequence of TLC, we could confirm that these four samples were astaxanthin.
Not only astaxanthin but also another spots were produced at II.
We regarded them as intermediary metabolites between zeaxanthin and astaxanthin.
The activity of pBad/araC promoter is measured in detail on another page. →click
acetone extract and vacuum concentration →protocol
Reference
[1] Yousry M. Antioxidant Activities of Astaxanthin and Related Carotenoids. J. Agric. Food Chem. 2000, 48, 1150-1154 [2]N.M. Sachindra, et al. Carotenoids in crabs from marine and fresh waters of India. LWT 38 (2005) 221–225
[3]MISAWA N, et al. Structure and Functional Analysis of a Marine Bacterial Carotenoid Biosynthesis Gene Cluster and Astaxanthin Biosynthetic Pathway Proposed at the Gene Level. JOURNAL OF BACTERIOLOGY, Nov. 1995, p. 6575–6584 Vol. 177, No. 22
[4]Seon-Kang C, et al. Characterization of bacterial β-carotene Ketolases, CrtW, from Mrine Bacteria by Complementation Analysis in Escherichia coli. MARINE BIOTECHNOLOGY(2005) 7, 515-522
[5]Seon-Kang C, et al. Characterization of bacterial β-carotene 3,3′-hydroxylases, CrtZ, and P450 in astaxanthin biosynthetic pathway and adonirubin production by gene combination in Escherichia coli. Appl Microbiol Biotechnol (2006) 72
[6]Nishizaki T, et al. Metabolic engineering of carotenoid biosynthesis in Escherichia coli by ordered gene assembly in Bacillus subtilis. Appl Environ Microbiol. 2007 Feb
[7]Arunkumar K, et al. Structure of a carotenoid gene cluster from Pantoea sp. strain C1B1Y and characterization of β-carotene hydroxylase(crtZ)gene by functional complementation in Escherichia coli. Carotenoid Science Vol.11 (2007)
appendix
β-carotene synthesis
Our team synthesized β-carotene under several conditions to confirm the activity of BioBrick parts designed by Cambredge 2009.
We used crtEBIY (BBa_K274210) to synthesize β-carotene. We compared the plasmids containing pSB3K3 (low-copy vector) with the plasmids containing pSB1A2 (high-copy vector) from the production. We also introduced the low-copy plasmid into E. Coli strain MG1655, JM109 and DH5α respectively.
We found that MG1655 and JM109 produce more β-carotene than DH5α in liquid culture and that MG1655 grow better than JM109 on plate culture. These results indicated that MG1655 is the best of the three strains to treat. For this reason, we decided to use only MG1655 to extract pigment.
We also made special competent cell, MG1655 and JM109, having a plasmid crtEBIY; pSB1A2 or crtEBIY; pSB3K3) for zeaxanthin and astaxanthin synthesis.(→S1, S2)
~acetone extract~
→protocol
Materials and Methods
The work flow to TLC from culture[3],[4]
Material
E. coli : strain MG1655
reagent : acetone, methanol, chloroform, hexane
TLC plate : silicagel 60F254
culture
1. The cells were cultured over night.
2. 50ul bacterial solution was taken from culture prepared in 1 and moved into 50ml LB culture with appropriate antibiotic. Inducer was added, if necessary.
3. The cells were incubated at 37°C, 24hour.
gathering E. coli and extract pigment
4. After the O.D reach 2.0, culture was centrifuged for 10minutes at 4°C, 6000rpm,10min
5. Supernatant was removed by decanting. The remainder was added with 1 ml ddH20 and vortexed.
6. Moved into 2ml tube by using P1000 pipetter at least twice.
7. Solution is centrifuged for 20minutes at 4°C, 14000rpm
8. Supernatant was removed completely using pipetter. About 100ul of pellet is obtained.
9. Pellet is added with 500ul acetone, and vortexed for 10 minutes to extract the pigment.
10. Solution is centrifuged for 20minutes at 4°C, 14000rpm
11. Acetone solution was replaced into 2 ml tube.
vacuum and concentrate
12. Acetone was removed using rotary evaporator.
13. 50 ul of Chloroform/Methanol(weight ratio of 9:1) solution was added.
14. Sample was vortexed and spinned down.
15. The water layer was removed.
TLC
16. Spot the sample onto TLC silica gel plate, and developed by acetone/hexane(weight ratio of 1:3) solution.
17. Spot was observed under visible light. "
"