Team:Tokyo Tech/Project/Apple Reporter/Results
From 2010.igem.org
(New page: {{Tokyo_Tech_Template2}} <div id="super_main_wrapper"> <div id="tf_menu"> <center> {| style="color:black; background-color:##ede8e2;" cellpadding="5" cellspacing="0" border="1" |+ <fo...) |
|||
| Line 29: | Line 29: | ||
<div id="tf_SubWrapper"> | <div id="tf_SubWrapper"> | ||
<!-- ここから始まるよ--> | <!-- ここから始まるよ--> | ||
| + | ==Results== | ||
| + | |||
| + | ===β-carotene synthesis=== | ||
| + | [[Image:tokyotech_Fig. 1.1.2.1. β-carotene.jpg|right|200px|Fig. 1.1.2. β-carotene]] | ||
| + | |||
| + | [http://en.wikipedia.org/wiki/Beta-Carotene →more information about β-carotene] | ||
| + | |||
| + | Our team synthesized β-carotene under several conditions to confirm the activity of BioBrick parts designed by Cambredge 2009. ([https://2010.igem.org/Team:Tokyo_Tech/Project/Apple_Reporter/Results See more...]) | ||
| + | |||
| + | We used ''crtEBIY'' (BBa_K274210) to synthesize β-carotene. We compared the plasmids containing pSB3K3 (low-copy vector) with the plasmids containing pSB1A2 (high-copy vector) from the production. We also introduced the low-copy plasmid into ''E. Coli'' strain MG1655, JM109 and DH5α respectively. | ||
| + | |||
| + | We found that MG1655 and JM109 produce more β-carotene than DH5α in liquid culture and that MG1655 grow better than JM109 on plate culture. These results indicated that MG1655 is the best of the three strains to treat. For this reason, we decided to use only MG1655 to extract pigment. | ||
| + | |||
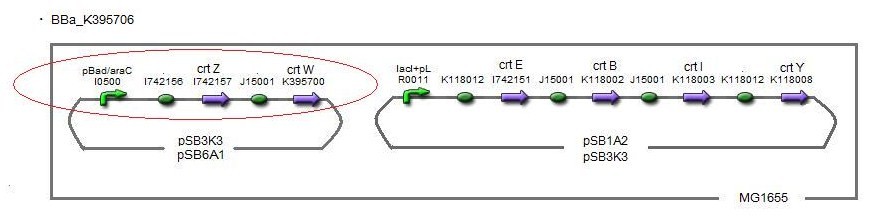
| + | We also made special competent cell, MG1655 and JM109, having a plasmid ''crtEBIY''; pSB1A2 or ''crtEBIY''; pSB3K3) for zeaxanthin and astaxanthin synthesis.(→[https://2010.igem.org/Image:Tokyotech_Fig._1.1.7.1.BBa_K395704.jpg figure1], [https://2010.igem.org/Image:Tokyotech_Fig._2-1-10_1BBa_K395706.jpg figure2]) | ||
| + | |||
| + | |||
| + | ~acetone extract~ | ||
| + | [[→protocol]] | ||
| + | |||
| + | [[Image:tokyotech_Fig. 1.1.3. β-carotene extract.jpg|300px|left|thumb|Fig. 2-1-3. β-carotene extract]] | ||
| + | |||
| + | |||
| + | [[Image:tokyotech_Fig. 1.1.4. β-carotene plate.jpg|300px|right|thumb|Fig. 2-1-4. β-carotene plate]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | [[Image:tokyotech_tokyotech_Fig. 1.1.5. note.jpg|500px|center|tokyotech_Fig. 1.1.5. note]] | ||
| + | |||
| + | ===zeaxanthin synthesis=== | ||
| + | [[Image:tokyotech_Fig. 1.1.6. zeaxanthin.jpg|right|200px|Fig. 1.1.6. zeaxanthin]] | ||
| + | |||
| + | [[→more information about zeaxanthin]] | ||
| + | |||
| + | Our team synthesized zeaxanthin by several kinds of construct to confirm the activity of BioBrick parts designed by Cambredge 2009 and Edinburgh 2007. By assembling the ''crtZ'', ''crtEBIY'' and appropriate promoters, we constructed new BioBrick parts to synthesize zeaxanthin and compared their function . | ||
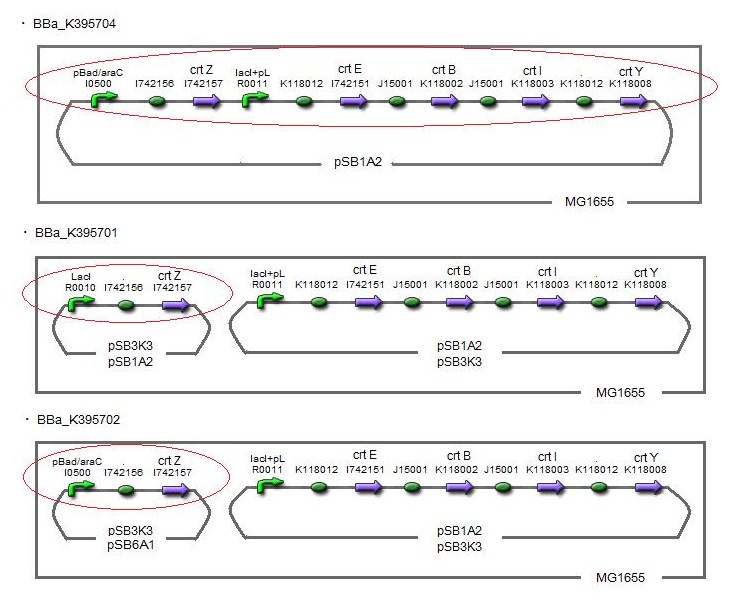
| + | (→[http://partsregistry.org/wiki/index.php?title=Part:BBa_K395701 BBa_K395701], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K395702 BBa_K395702], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K395704 BBa_K395704]) | ||
| + | We constructed two types of zeaxanthin synthetic construct. ([https://2010.igem.org/Team:Tokyo_Tech/Project/Apple_Reporter/Results See more...]) | ||
| + | |||
| + | One is that ''crtZ''(BBa_I742158) and ''crtEBIY''(BBa_K274210) were assembled on a single plasmid.(We call this construct “single plasmid construct”.) | ||
| + | The other is that ''crtZ'' and ''crtEBIY''were assembled on different plasmids.(we call this type of constructs “double plasmids construct”.) | ||
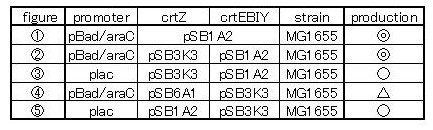
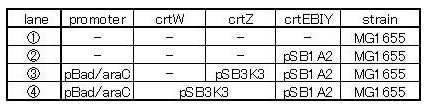
| + | We made 5 combinations shown in table<1>. | ||
| + | |||
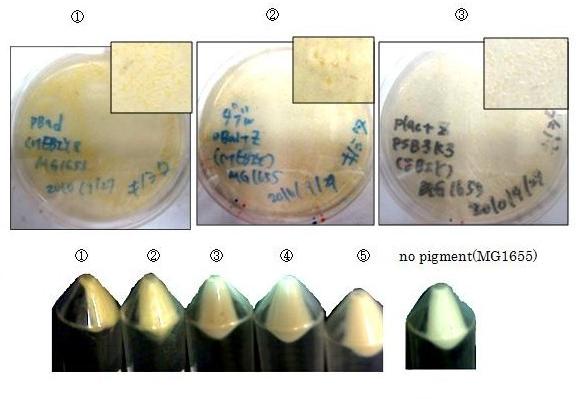
| + | Constructs containing pBad/araC promoter on high-copy vector show the best pruduction and double plasmids construct shows as much pruduction as single plasmid construct under same promoter conbination. Construct with pBad/araC promoter shows better production than that with plac promoter. Construct that ''crtEBIY'' on high-copy vector and ''crtZ'' on low-copy vector shows as much expression as construct that ''crtEBIY'' on low-copy vector and ''crtZ'' on high-copy vector.(Actually, ③ showed a little more production than ⑤.) | ||
| + | |||
| + | |||
| + | |||
| + | [[Image:tokyotech_Fig. 1.1.7.1.BBa_K395704.jpg|right|320px|thumb|S1]] | ||
| + | |||
| + | |||
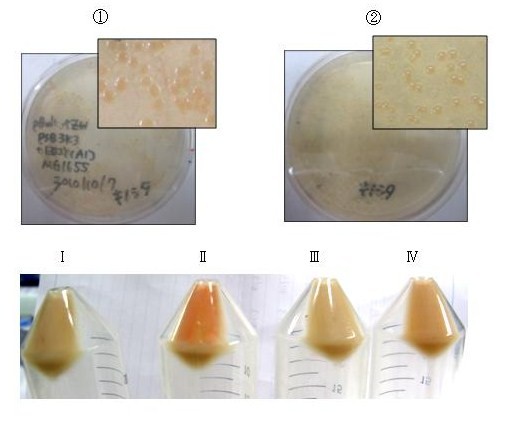
| + | [[Image:tokyotech_Fig. 1.1.8.1. zeaxanthin pellet and plate.jpg|left|290px|Fig. 1.1.8.1. zeaxanthin pellet and plate]] | ||
| + | |||
| + | |||
| + | [[Image:tokyotech_table. 1.1.1.2.jpg|500px|center|thumb|table<1>]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | From the result, we decided to use pBad/araC promoter and double plasmids construct. | ||
| + | |||
| + | ===astaxanthin synthesis=== | ||
| + | [[Image:tokyotech_Fig. 1.1.9. zeaxanthin.jpg|right|200px|Fig. 1.1.9. zeaxanthin]] | ||
| + | |||
| + | [[→more information about astaxanthin]] | ||
| + | |||
| + | Our team synthesized astaxanthin to make the pathway reach the end by assembling ''crtW'' and zeaxanthin synthesis constructs. After constructing new BioBrick parts to synthesize astaxanthin we compared their function. (→[http://partsregistry.org/wiki/index.php?title=Part:BBa_K395706 BBa_K395706]), We also examine the production controlled by activity of pBad/araC promoter. ([https://2010.igem.org/Team:Tokyo_Tech/Project/Apple_Reporter/Results See more...]) | ||
| + | |||
| + | |||
| + | The activity of this promoter is measured in detail on another page. [[→click]] | ||
| + | |||
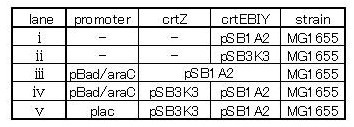
| + | We made 4 combinations shown in table<2> | ||
| + | |||
| + | |||
| + | [[Image:tokyotech_Fig. 2-1-10 1BBa_K395706.jpg|center|500px|thumb|S2]] | ||
| + | |||
| + | |||
| + | [[Image:tokyotech_Fig. 2-1-11 astaxanthin pellet and plate.jpg|left|310px|Fig. 2-1-11 astaxanthin pellet and plate]] | ||
| + | |||
| + | ~acetone extract and vacuum concentration~ [[→protocol]] | ||
| + | [[Image:tokyotech_Fig. 2-1-12 astaxanthin extract.jpg|right|310px|Fig. 2-1-12 astaxanthin extract]] | ||
| + | |||
| + | |||
| + | [[Image:tokyotech_table. 2.1.3.jpg|500px|center|thumb|table<2>]] | ||
| + | |||
| + | ===Separation of carotenoids with TLC=== | ||
| + | |||
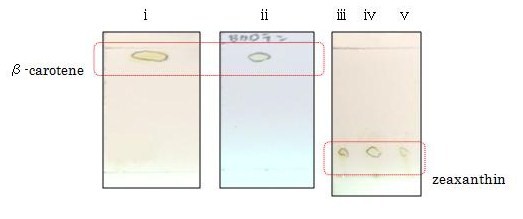
| + | We performed thin layer chromatography (TLC) to confirm the products of β-carotene, zeaxanthin, astaxanthin were synthesized. ([https://2010.igem.org/Team:Tokyo_Tech/Project/Apple_Reporter/Results See more...]) | ||
| + | |||
| + | The lanes are ①control(FPP), ②β-carotene, ③zeaxanthin, ④astaxanthin from the left. We can tell ②,③ and ④ are yellow-orange, golden-yellow and red-orange pigment, respectively. These results are consistent with the Rf and color of β-carotene, zeaxanthin, astaxanthin respectively and indicated that target products were synthesized as expected. [Reference: Carotenoids in crabs from marine and fresh waters of India ] | ||
| + | |||
| + | [[Image:tokyotech_table. 2-1-3.jpg|400px|center|table<3>]] | ||
| + | |||
| + | |||
| + | [[Image:tokyotech_Fig. 2-1-13 allTLC.jpg|500px|center|Fig. 2-1-13 allTLC]] | ||
| + | |||
| + | β-carotene and zeaxanthin | ||
| + | |||
| + | [[Image:tokyotech_table. 2-1-4.jpg|315px|left|table<4>]] | ||
| + | |||
| + | [[Image:tokyotech_Fig. 2-1-14 TLC β-caroten zeaxanthin.jpg|315px|right|Fig. 2-1-14 TLC β-caroten zeaxanthin]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
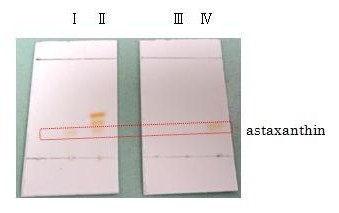
| + | astaxanthin | ||
| + | [[Image:tokyotech_table 2-1-5-1.jpg|320px|left|table<5>]] | ||
| + | |||
| + | [[Image:tokyotech_Fig. 2-1-15 TLC astaxanthin.jpg|300px|right|Fig. 2-1-15 TLC astaxanthin]] | ||
| + | |||
| + | Not only astaxanthin but also another pigments were priduced at Ⅱ. | ||
| + | |||
| + | We regarded them as intermediary metabolites between zeaxanthin and astaxanthin. | ||
<!-- ここまでね--> | <!-- ここまでね--> | ||
Revision as of 09:37, 27 October 2010
Contents |
Results
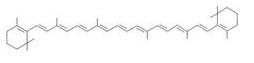
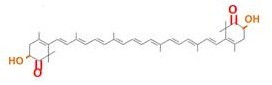
β-carotene synthesis
[http://en.wikipedia.org/wiki/Beta-Carotene →more information about β-carotene]
Our team synthesized β-carotene under several conditions to confirm the activity of BioBrick parts designed by Cambredge 2009. (See more...)
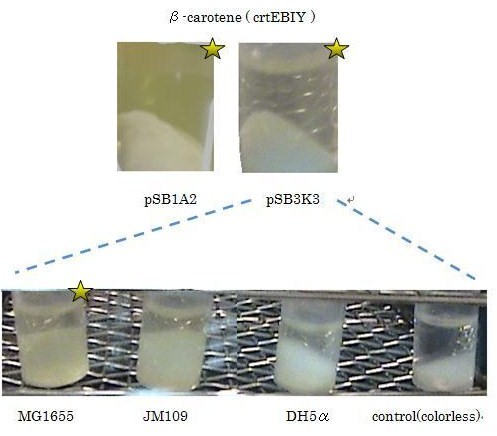
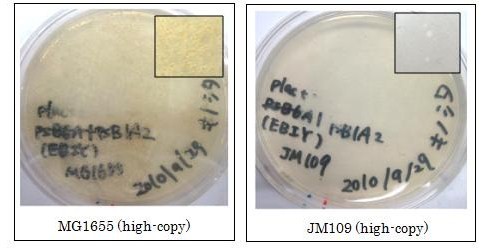
We used crtEBIY (BBa_K274210) to synthesize β-carotene. We compared the plasmids containing pSB3K3 (low-copy vector) with the plasmids containing pSB1A2 (high-copy vector) from the production. We also introduced the low-copy plasmid into E. Coli strain MG1655, JM109 and DH5α respectively.
We found that MG1655 and JM109 produce more β-carotene than DH5α in liquid culture and that MG1655 grow better than JM109 on plate culture. These results indicated that MG1655 is the best of the three strains to treat. For this reason, we decided to use only MG1655 to extract pigment.
We also made special competent cell, MG1655 and JM109, having a plasmid crtEBIY; pSB1A2 or crtEBIY; pSB3K3) for zeaxanthin and astaxanthin synthesis.(→figure1, figure2)
~acetone extract~
→protocol
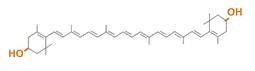
zeaxanthin synthesis
→more information about zeaxanthin
Our team synthesized zeaxanthin by several kinds of construct to confirm the activity of BioBrick parts designed by Cambredge 2009 and Edinburgh 2007. By assembling the crtZ, crtEBIY and appropriate promoters, we constructed new BioBrick parts to synthesize zeaxanthin and compared their function . (→[http://partsregistry.org/wiki/index.php?title=Part:BBa_K395701 BBa_K395701], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K395702 BBa_K395702], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K395704 BBa_K395704]) We constructed two types of zeaxanthin synthetic construct. (See more...)
One is that crtZ(BBa_I742158) and crtEBIY(BBa_K274210) were assembled on a single plasmid.(We call this construct “single plasmid construct”.) The other is that crtZ and crtEBIYwere assembled on different plasmids.(we call this type of constructs “double plasmids construct”.) We made 5 combinations shown in table<1>.
Constructs containing pBad/araC promoter on high-copy vector show the best pruduction and double plasmids construct shows as much pruduction as single plasmid construct under same promoter conbination. Construct with pBad/araC promoter shows better production than that with plac promoter. Construct that crtEBIY on high-copy vector and crtZ on low-copy vector shows as much expression as construct that crtEBIY on low-copy vector and crtZ on high-copy vector.(Actually, ③ showed a little more production than ⑤.)
From the result, we decided to use pBad/araC promoter and double plasmids construct.
astaxanthin synthesis
→more information about astaxanthin
Our team synthesized astaxanthin to make the pathway reach the end by assembling crtW and zeaxanthin synthesis constructs. After constructing new BioBrick parts to synthesize astaxanthin we compared their function. (→[http://partsregistry.org/wiki/index.php?title=Part:BBa_K395706 BBa_K395706]), We also examine the production controlled by activity of pBad/araC promoter. (See more...)
The activity of this promoter is measured in detail on another page. →click
We made 4 combinations shown in table<2>
~acetone extract and vacuum concentration~ →protocol
Separation of carotenoids with TLC
We performed thin layer chromatography (TLC) to confirm the products of β-carotene, zeaxanthin, astaxanthin were synthesized. (See more...)
The lanes are ①control(FPP), ②β-carotene, ③zeaxanthin, ④astaxanthin from the left. We can tell ②,③ and ④ are yellow-orange, golden-yellow and red-orange pigment, respectively. These results are consistent with the Rf and color of β-carotene, zeaxanthin, astaxanthin respectively and indicated that target products were synthesized as expected. [Reference: Carotenoids in crabs from marine and fresh waters of India ]
β-carotene and zeaxanthin
astaxanthin
Not only astaxanthin but also another pigments were priduced at Ⅱ.
We regarded them as intermediary metabolites between zeaxanthin and astaxanthin.
 "
"