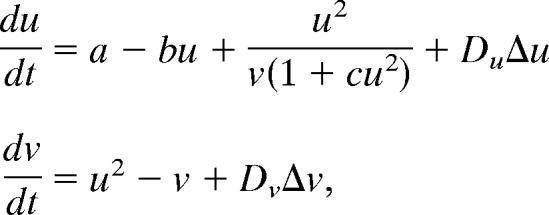Team:Tokyo Metropolitan/Project/Pattern
From 2010.igem.org

Contents |
Overall project
Reaction-Diffusion model
Reaction-diffusion equation proposed by A.Turing, 1952.Turing is the famous mathematician of England, as who invent “Turing machine”. This equation describe reaction and diffusion of two substances, and they form specific pattern autonomously. pattern which read by these substances are similar to animal specific skin pattern such as spot pattern of Panther, stripe pattern of zebra, and so on. But the relationship between equation and animal skin pattern was not clear for long days. in 1995,Kondo.S demonstrated that skin pattern of Pomacanthus can describe by using reaction-diffusion equation. Kondo our purpose is, to demonstrate existence of Turing pattern in organisms, by reproduce patterns using E.coli. and verify the theory.
approach
We described that Reaction-Diffusion equation shows reaction and diffusion of “substances”. To form patterns, these “substances” whould be important. We need morphogen, “Activator” and “Inhibitor” as substance. When these substance fulfill some conditions, patterns would reproduce such as spot patterns like panther, or stripe patterns like zebra and so on. this is already demonstrated in computer simulations.(References) We suppose that a bacteria as an individual cell. These “cells” express the genes which cording patterning substance. and reproduce autonomous patterns by their interact. We have some ideas to make these patterns more interesting. One is use of temperature-sensitive bacteria, to make different patterns by temperature in same culture. Another idea is use of multiple substances. Increasing types of substances, we expect that can make more complicated patterns.
the model
How we meet the condition?
We described that to reproduce the model, we need two substances, Activator and inhibitor. but we need more conditions about relationships between Activator and inhibitor;
1. There are two components- the activator and inhibitor
2.Activator's self activation
3.Inhibitor's activation by activator
4.activator's inhibition by inhibitor
5.diffusion speed: Inhibitor>Activator
by following these condition, we designed the original reaction-diffusion model. The first condition ”two components- the activator and inhibitor” is already cleared. We designed Activator and Inhibitor like these;
Activator
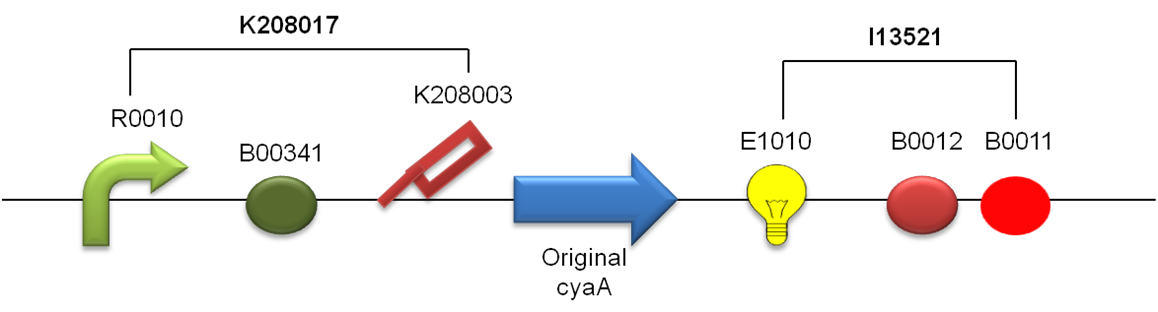
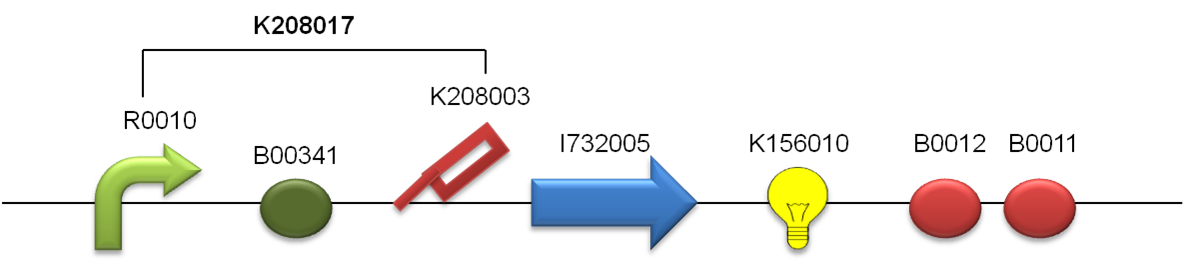
cAMP(cyclic adenosine monophosphate) is the second messenger of cellular signaling. cAMP activate global transcriptional factor CRP(cyclicAMP receptor protein). In our project, we introduce CyaA(cAMP synthase) to E.coli, and express cAMP. Then, activate CRP. This cascade fulfill first condition ,”Self-activation of Activator”. and cAMP secrete from activator cell and effect to Inhibitor cell . So, we can fulfill second condition, ”Activation of Inhibitor by Activator”. We designed biobrick as following. Putting CRP binding site, lac promoter, lacI binding site,(these sites become important to fulfill other conditions. We explain later.) RBS and ompA signal , on the upstream(5’ side) of Biobrick. these parts are Designed by USU iGEM2009,from team Utah state2009, for fused with a silver fusion compatible protein to make a functional device for secretion studies(http://partsregistry.org/Part:BBa_K208017). we use this parts for secretion of cyaA and cAMP. when cellular CRP bind to CRP binding site of this parts, transcription of cyaA, and mRFP1(and terminators) will start. We going to register (http://partsregistry.org/Part:BBa_K336060) as a new parts.
Inhibitor
We choose Inhibitor factor as Glucose. When lacZ(β-galactositase/Glucose synthase) express by CRP from Activator cell, synthase transport by ompA signal to periplasm. On periprasm, sythase synthesys Glucose. It effect to Activator cell’s lacI, and repress transcription of Activator plasmid. So, we can fulfill the 4th condition “Activator’s Inhibition by Inhibitor.” Biobrick of Inhibitor have also same structure with Activator. from Promoter to ompA signal using BBa_K208017 too. lacZ parts is taken from BBa_I732901 Designed by Zhan Jian, iGEM07_USTC, which containing full-length of lacZ(http://partsregistry.org/Part:BBa_I732901).
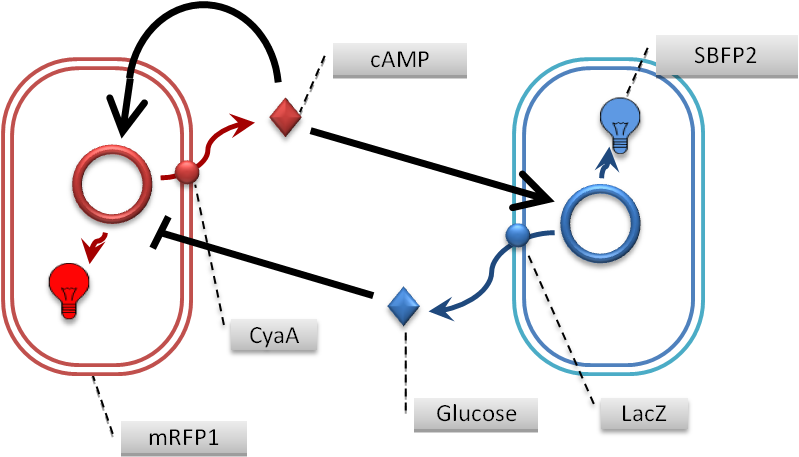
Relationship between activator and Inhibitor
We’d like to design the reaction-diffusion relationship by using famous mechanism of transcriptional regulations; Lac operon and cataborite repression. When cyaA transcripted by activator plasmid , cyaA transpoeted into periprasm layer by signaling signal sequence,ompA. After that,we leave permability of periprasmlayer. If the periprasm layer doesn’t transmit cAMP,which synthesized by cyaA, we going to leave life span of bacteria. If activator E.coli died, they going to explode. We expect the exprotion, which burst cyaA.After that, when cyaA explode onto medium, we expect which synthesize of CRP on the medium ,from cyaA when the CRP synthesized on the medium and incorporated into inhibitor E.coli, it going to activate the transcription of the Inhibitor, lacZ and the glucose. In activator cell, cyaA is transcripted by the Biobrick,and cyaA(transcriptional product)it transported by signal sequence into periprasm. After that, we leave the permability of periprasm.
In concretely, the activate of Inhibitor Biobrick, the lacZ,cording βgalactoristase is formed. βgalactositase also transported by signal sequence, and catabolite the lactose ,then form the glucose and lactose. By glucose absorpt 、it inhibit the transcription of Activate factor,cyaA and cAMP. And glucose going to consume.Mechanism going to be the Inhibit pathway or plasmid doesn’t have LacI binding site. So, Inihibitor cell doesn’to accept influence of the glucose.
In each plasmid,activator and Inihibitor,have the reportergene(BFP or RFP)on downstream of the plasmid. So, we can see the which constlact is activated. We can’t predict that how the pattern formatted by these relationships. So, we have to make predict the relationship based on the reaction-diffusion model between the Activator cells and Inhibitor cells.
now, we reached the production of the parts. We need to examine the pattern which made from our construct form the patten or not,and reasons. we going to report out result on jamboree and poster settion. Please comfirm the answer when you come to our space! :D
Acknowledgment
We whould like to thank; NIPPON GENE CO.,LTD,Merck & Co.,Inc.,Leave a Nest Co.,Ltd.,The Consortium of Universities in Hachioji,Sanplatec corp.,SARSTEDT Inc., NK system(Nihon ika kikai seisaku-sho), Pfizer Asia Research Inc. Mr.Shinji Ogawa, and Cosmotec Co.,Ltd.Mr.Yukio Takamizawa for providing us instrument and reagent. Dr.K.Tamura, Dr.H.Saiga,Dr.H.Kawahara,Dr.N.Kachi, Dr.Y.Okabe, Dr.Y.Murakami,and Mr.Y.Tsuda for many advices and cooperation to us. Member of the Evolutionaly genetics laboratory for let us the spaces to do the experiments. Member of iGEM Osaka 2009 for instructing the method of medium. and Mr.Yuusuke Mii for providing us the RD simuration soft.
References
- Kondo S. et al. , “Theoretical analysis of mechanisms that generate the pigmentation pattern of animals.” Cell Dev Biol. 20(1):82-9.(2009)
- Kondo S. et al., ”Interactions between zebrafish pigment cells responsible for the generation of Turning patterns.” Proc Natl Acad Sci U S A. 106(21):8429-34.(2009)
- Horikawa K., Ishimatsu K., Yoshimoto E., Kondo S.,Takeda H. “Noise-resistant and synchronized oscillation of the segmentation clock” Nature 441 (7094) :719-723 (2006)
- Kondo S. “The reaction-diffusion system: a mechanism for autonomous pattern formation in the animal skin.” Genes to Cells 7 (6): 535-541.(2002)
- Kondo S. and Asai R. “A reaction-diffusion wave on the skin of the marine angelfish Pomacanthus.” Nature 376 (6543) :765-768 (1995)
- S. sick, S. timmer, T. schlake “WNT and DKK hair follicle spacing through a reaction-diffusion mechanism” Science vol.314 1447-1450(2006)
- Turing AM, "The Chemical Basis of Morphogenesis", Philosophical Transactions of The Royal Society of London, series B, 237:37–72, 1952.
- Gierer A and Meinhardt H, "A theory of biologycal pattern formation", Kybernetik 12:30-39, 1972
- S.Kondo and T.Miura “Reaction-Diffusion Model as a Framework for Understanding Biological Pattern Formation” Science vol.329 1616-1620(2010)
 "
"