Team:Washington/Gram Positive/Test
From 2010.igem.org
(→Enzyme Assay) |
(→Characterization of best hits) |
||
| (163 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | __NOTOC__ | ||
{{Template:Team:Washington/Templates/Header}} | {{Template:Team:Washington/Templates/Header}} | ||
<html> | <html> | ||
| Line 24: | Line 25: | ||
</html> | </html> | ||
<!---------------------------------------PAGE CONTENT GOES BELOW THIS----------------------------------------> | <!---------------------------------------PAGE CONTENT GOES BELOW THIS----------------------------------------> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | == | + | =Construction of CapD_CP= |
| + | ==CapD_CP runs on a gel as one clean band== | ||
| - | |||
| - | + | [[Image:CapDandCapDCPGell.jpg|480px|left|thumb|'''Figure 1. CapD_CP as expected. Right: CapD has ambiguous bands, making quantifying difficult and assaying protein activity less accurate | |
| - | + | ''']] | |
| - | |||
| - | |||
| - | + | Depicted in Figure 1 is a gel comparing purified CapD and CapD_CP. This demonstrates one of the major advantages of CapD_CP,in addition to the fact that it is easy to express, it comes out in one clean band and is easy to quantify. | |
| - | + | ||
| - | + | ||
| - | |||
| - | + | CapD shows three bands, while the lower two bands are the expected pieces that occur after the enzyme undergoes self cleavage, the upper band is of unclear origin. Its mass corresponds to what would either be the unprocessed, inactive and monomeric form of the enzymes or the dimeric form that didn't denature. This ambiguity makes the amount of active CapD enzyme difficult to quantify. | |
| - | |||
| - | |||
| - | <br | + | <br style="clear: both" /> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | ==CapD_CP is expressed at EXACTLY the correct molecular weight== |
| - | + | In Figure 2 (below) we show the mass spec conformation of the molecular weight of CapD_CP. As expected, the N-terminal methionine is cleaved off by the native E. coli methionine aminopeptidase, resulting in a high level of pure active enzyme. | |
| - | + | ||
| - | + | ||
| - | + | [[Image:CapDCP_MassSpecNoMethionine.png|600px|left|thumb|''' Figure 2. Expected weight of CapD_CP without Methionine=55285Da, with Methionine=55417Da. Our mass spec detected a peak at 55274.8Da (no Methionine) well within the 0.02% error limit for our mass spec''']] | |
| + | <br style="clear: both" /> | ||
| - | + | ==The circular permutation didn't result in a loss of activity== | |
| - | |||
| - | |||
| - | |||
| - | + | In the table 1 (below), we show the kinetic paramters for CapD and CapD_CP as determined using our [https://2010.igem.org/Team:Washington/Protocols/EnzymeAssayCapD| enzyme assay]. | |
| - | + | Due to the heterogenous mix of active (processed) and inactive (unprocessed) protein, the kcat is difficult to quantify. The data we obtained suggests that the kinetic parameters of CapD and CapD_CP are within error of each other. Therefore the circular permutation of CapD did not have a negative effect on catalytic activity, suggesting that auto-processing is not required for catalysis but simply a regulatory feature. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ==''' | + | [[Image:Washington_CP_D_Constants.png|thumb|500px|center| '''Table 1. Kinetic properties determined for a Michaelis-Menten profile of CapD and CapD_CP, as described in our protocols section.''']] |
| - | By standardizing the activity slope of each design relative to CapD_CP, a scatterplot easily portrays the qualities of each mutant. Several designs show negative catalytic curves similar to the catalytic knockouts. Some immediately show a negative activity curve meaning decrease in transpeptidation, hydrolysis, or both. | + | |
| + | |||
| + | <br style="clear: both" /> | ||
| + | |||
| + | ==Catalytic Residue knock-outs show that CapD_CP is catalyzing the reaction as expected== | ||
| + | Before we could predict which mutations increase hydrolysis capability, we needed to validate that the circularly permuted version of CapD had measurable activity for further assessments. We also hypothesized a threonine residue in the catalytic site of CapD_CP plays an important role in the catalysis reaction and mutating it will eliminate all enzymatic activity. Thus using [https://2010.igem.org/Team:Washington/Project/Tools/FoldIt FoldIt ]we created two mutants, T2V and T2A, to act as negative controls and ran an [https://2010.igem.org/Team:Washington/Protocols/EnzymeAssayCapD enzyme assay] to confirm our hypothesis. The result shown in figure 3 (below) of this assay confirmed our hypothesis that CapD_CP has enzymatic activity compared to the two catalytic knockouts. The relatively flat activity curves of the knockout mutants confirmed the hypothesis of the threonine's role in the catalytic site. | ||
| + | |||
| + | [[Image:Washington_Confirming_CP_activity_revised7.jpg|thumb|750px|left|'''Figure 3: Confirming the activity of CapD_CP by comparing it to two CapD_CP knockouts, T2A and T2V.''']] | ||
| + | <br style="clear: both" /> | ||
| + | |||
| + | =Removing CapD_CP Transpeptidase Activity= | ||
| + | <span id = mutant> | ||
| + | ==Screening Mutant Libraries== | ||
| + | By standardizing the activity slope of each design relative to CapD_CP, a scatterplot easily portrays the qualities of each mutant. Several designs show negative catalytic curves similar to the catalytic knockouts. Some immediately show a negative activity curve meaning decrease in transpeptidation, hydrolysis, or both. F24H is our most promising mutant hydrolase design. | ||
| + | |||
| + | [[Image:Washington_Mutant_design.jpg|thumb|750px|center|'''Figure 4. Plot of the Transpeptidation vs. Hydrolysis, relative to the starting protein CapD_CP. The goal is to remove as much transpeptidation activity as possible while maintaining or increasing hydrolysis activity. This will allow the protein to break down (via the hydrolysis reaction) the protective coat without building it back up (via the transpeptidation reaction)''']] | ||
| + | |||
| + | [[Team:Washington/Gram_Positive/Test/MutantDataTable|Mutant activity relative to CapD_CP in table form]] | ||
| + | <br style="clear: both" /> | ||
| + | |||
| + | <span id = capH> | ||
| + | |||
| + | ==Characterization of best hits== | ||
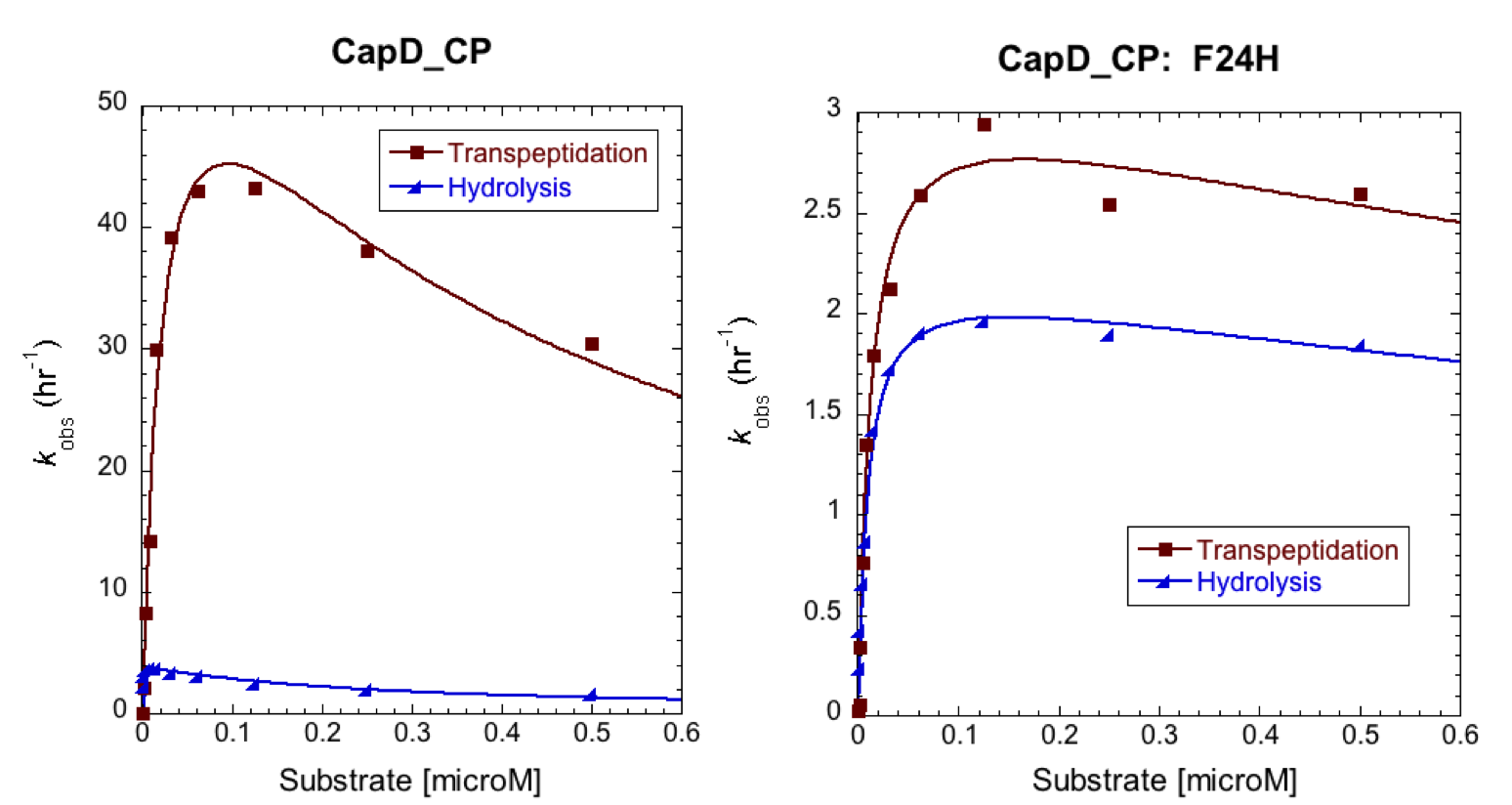
| + | To further characterize the most promising mutant, F24H, we ran a Michaelis-Menten profile in order to obtain it's kinetic parameters and compare them to CapD. In this case we ran our standard enzyme assay (5nM enzyme), but we varied the substrate from 1microM down to 0microM. The resulting substrate vs. velocity curves for CapD_CP and the F24H variant are shown below. Both enzymes showed canonical substrate inhibition trends, so the curve we fit them to was: ''k''obs = (''k''cat * S0)/(Km+S0*(1+S0/Ki)). | ||
| + | |||
| + | [[Image:Washington_CP_F24H_MMCurves.png|thumb|700px|center|'''Figure 5. Substrate vs. Velocity curves to determine the catalytic paramters if CapD_CP (left) and CapD_CP-F24H (right).''']] | ||
| + | |||
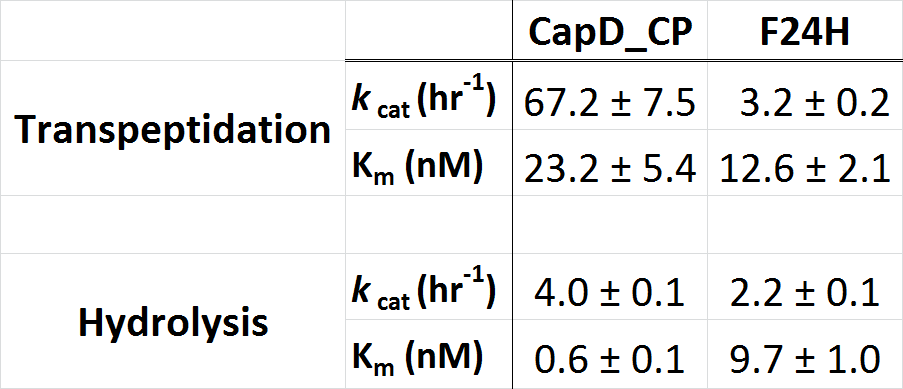
| + | [[Image:Washington_CP_F24H_Constants.png|thumb|400px|center|'''Table 2. Kinetic parameters of CapD_CP vs. CapD_CP-F24H show that the mutant achieved the desired goal of having a minimal effect on hydrolysis activity (''k''cat), but has significantly reduced transpeptidation activity.''']] | ||
| + | |||
| + | <br style="clear: both" /> | ||
| + | </span> | ||
| + | |||
| + | ==Summary== | ||
| + | |||
| + | These results show that our mutant has converted CapD_CP from an enzyme that strongly prefers transpeptidation into one that performs transpeptidation equally as well as it does hydrolysis. The effects of this on anthrax need to be more thoroughly characterized, but that is beyond the scope of our project. In addition, we hope to continue working on this project after the completion of iGEM in order to find additional variants that will start increasing hydrolysis activity. | ||
| + | |||
| + | |||
| + | <br style="clear: both" /> | ||
| - | |||
<!---------------------------------------PAGE CONTENT GOES ABOVE THIS----------------------------------------> | <!---------------------------------------PAGE CONTENT GOES ABOVE THIS----------------------------------------> | ||
<div style="text-align:center"> | <div style="text-align:center"> | ||
| - | '''← [[Team:Washington/ | + | '''← [[Team:Washington/Gram Positive/Build|Building the Gram(+) Therapeutic]]''' |
| | ||
| | ||
| | ||
| - | '''[[Team:Washington/ | + | '''[[Team:Washington/Tools Created|Overview of Tools We Created]] →''' |
| - | + | ||
{{Template:Team:Washington/Templates/Footer}} | {{Template:Team:Washington/Templates/Footer}} | ||
Latest revision as of 20:59, 27 October 2010
Construction of CapD_CP
CapD_CP runs on a gel as one clean band
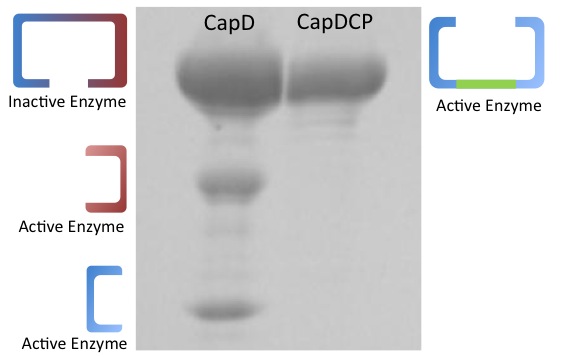
Depicted in Figure 1 is a gel comparing purified CapD and CapD_CP. This demonstrates one of the major advantages of CapD_CP,in addition to the fact that it is easy to express, it comes out in one clean band and is easy to quantify.
CapD shows three bands, while the lower two bands are the expected pieces that occur after the enzyme undergoes self cleavage, the upper band is of unclear origin. Its mass corresponds to what would either be the unprocessed, inactive and monomeric form of the enzymes or the dimeric form that didn't denature. This ambiguity makes the amount of active CapD enzyme difficult to quantify.
CapD_CP is expressed at EXACTLY the correct molecular weight
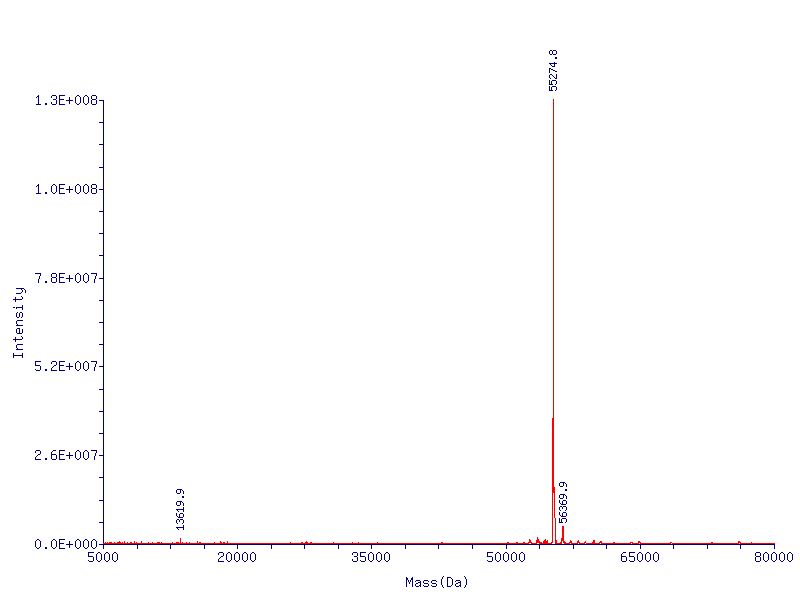
In Figure 2 (below) we show the mass spec conformation of the molecular weight of CapD_CP. As expected, the N-terminal methionine is cleaved off by the native E. coli methionine aminopeptidase, resulting in a high level of pure active enzyme.
The circular permutation didn't result in a loss of activity
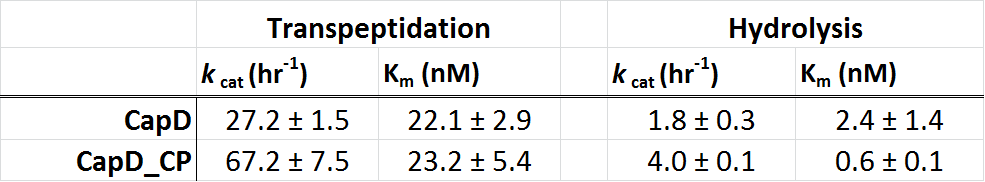
In the table 1 (below), we show the kinetic paramters for CapD and CapD_CP as determined using our enzyme assay. Due to the heterogenous mix of active (processed) and inactive (unprocessed) protein, the kcat is difficult to quantify. The data we obtained suggests that the kinetic parameters of CapD and CapD_CP are within error of each other. Therefore the circular permutation of CapD did not have a negative effect on catalytic activity, suggesting that auto-processing is not required for catalysis but simply a regulatory feature.
Catalytic Residue knock-outs show that CapD_CP is catalyzing the reaction as expected
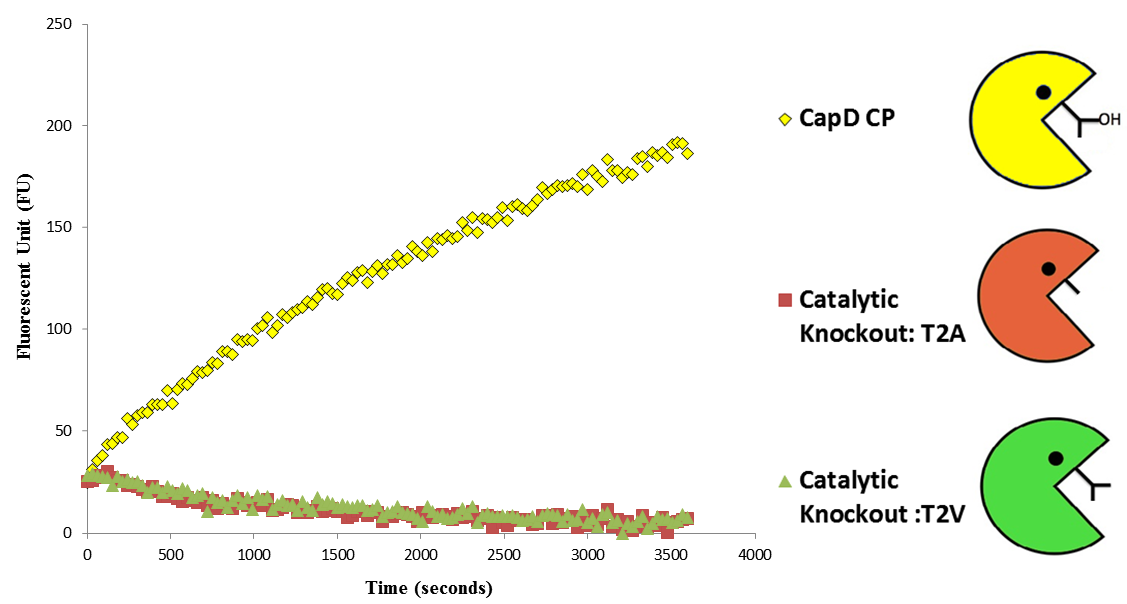
Before we could predict which mutations increase hydrolysis capability, we needed to validate that the circularly permuted version of CapD had measurable activity for further assessments. We also hypothesized a threonine residue in the catalytic site of CapD_CP plays an important role in the catalysis reaction and mutating it will eliminate all enzymatic activity. Thus using FoldIt we created two mutants, T2V and T2A, to act as negative controls and ran an enzyme assay to confirm our hypothesis. The result shown in figure 3 (below) of this assay confirmed our hypothesis that CapD_CP has enzymatic activity compared to the two catalytic knockouts. The relatively flat activity curves of the knockout mutants confirmed the hypothesis of the threonine's role in the catalytic site.
Removing CapD_CP Transpeptidase Activity
Screening Mutant Libraries
By standardizing the activity slope of each design relative to CapD_CP, a scatterplot easily portrays the qualities of each mutant. Several designs show negative catalytic curves similar to the catalytic knockouts. Some immediately show a negative activity curve meaning decrease in transpeptidation, hydrolysis, or both. F24H is our most promising mutant hydrolase design.
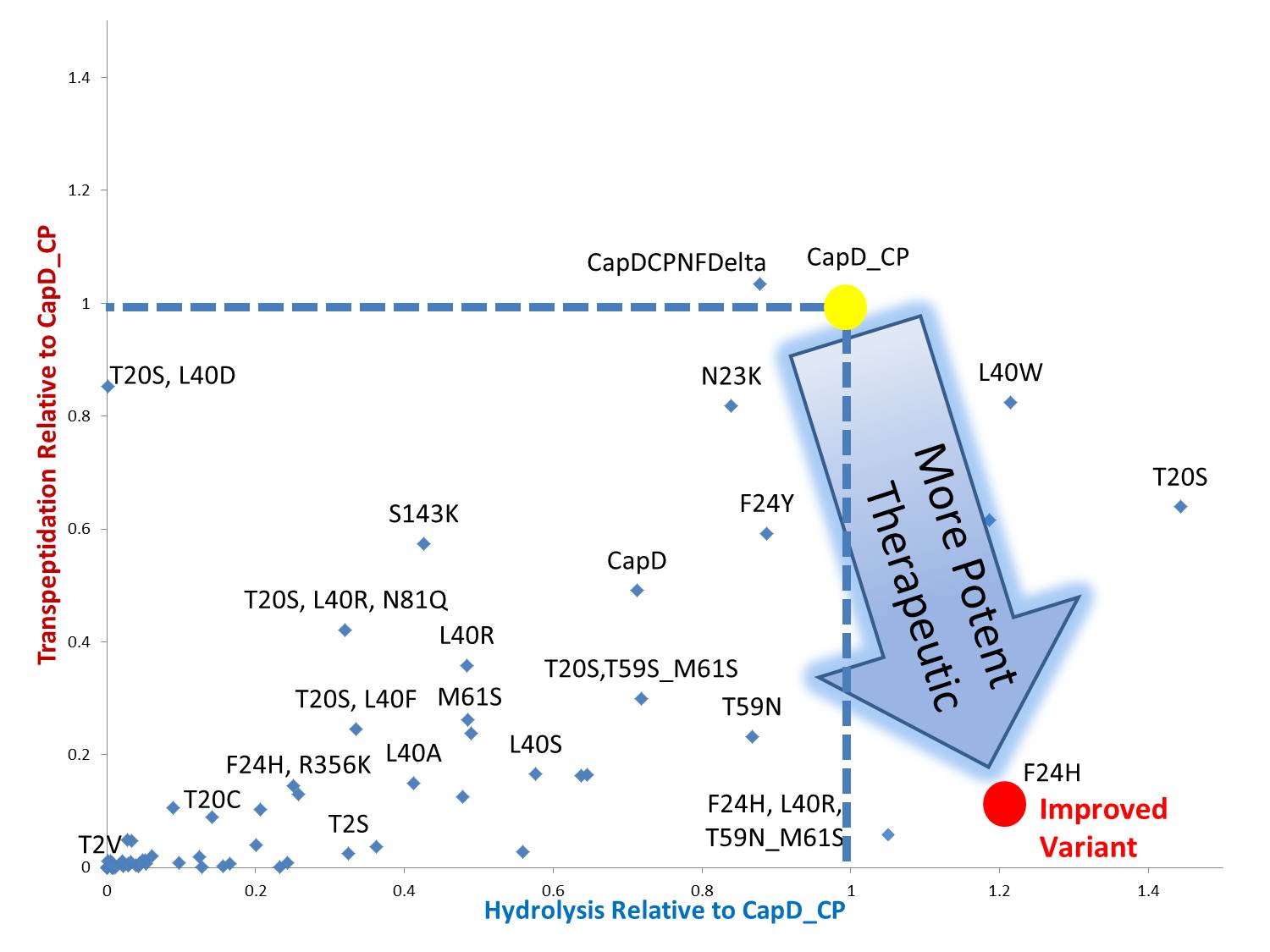
Mutant activity relative to CapD_CP in table form
Characterization of best hits
To further characterize the most promising mutant, F24H, we ran a Michaelis-Menten profile in order to obtain it's kinetic parameters and compare them to CapD. In this case we ran our standard enzyme assay (5nM enzyme), but we varied the substrate from 1microM down to 0microM. The resulting substrate vs. velocity curves for CapD_CP and the F24H variant are shown below. Both enzymes showed canonical substrate inhibition trends, so the curve we fit them to was: kobs = (kcat * S0)/(Km+S0*(1+S0/Ki)).
Summary
These results show that our mutant has converted CapD_CP from an enzyme that strongly prefers transpeptidation into one that performs transpeptidation equally as well as it does hydrolysis. The effects of this on anthrax need to be more thoroughly characterized, but that is beyond the scope of our project. In addition, we hope to continue working on this project after the completion of iGEM in order to find additional variants that will start increasing hydrolysis activity.
 "
"