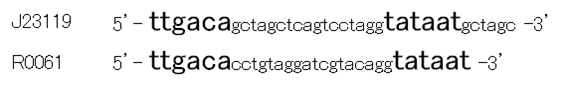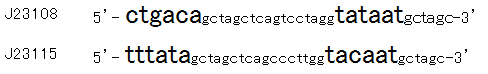Team:Tokyo Tech/Project/Artificial Cooperation System/lux act rep
From 2010.igem.org
Contents
|
lux activation/repression part
I, characterization of R0061 (promoter repressed by LuxR/3OC6HSL)
Introduction
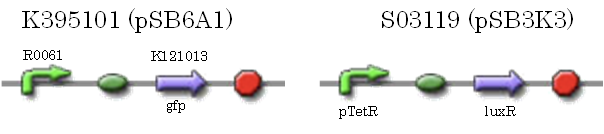
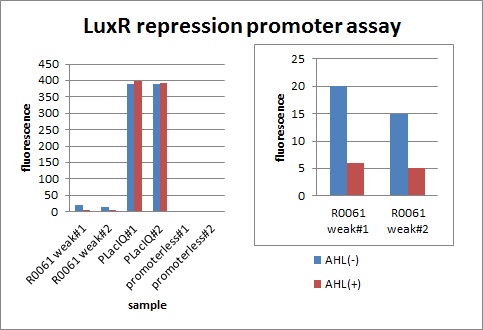
We characterized luxR repression promoter. In the Artificial Cooperation System, we inserted chloramphenicol resistance coding sequence into this promoter. Thus, this promoter plays an important role in Artificial Cooperation System. We wanted to characterize the strength of this promoter which has never been done before in BioBrick in order to design new promoter based on this data. First, we confirmed R0061, which is a existing BioBrick promoter repressed by LuxR/3OC6HSL complex. To confirm this promoter, we constructed following two plasmids (fig〇〇and fig〇〇)
We introduced these two plasmid into DH5α.
Result
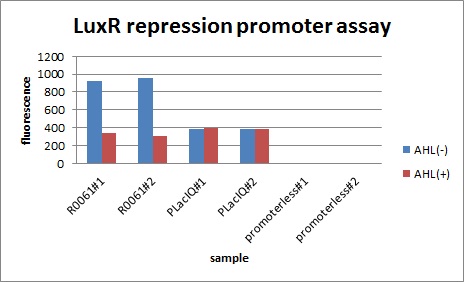
In the presence of 3OC6HSL(100nM), the fluorescence is lower than in the absence of 3OC6HSL. We used a fusion of placIQ (I14032) to gfp (K121013) as a positive control and used promoterless gfp (K121013) as a negative control. We measured fluorescence by flow cytometry 3 hour after addition of 100nM 3OC6HSL.
Conclution
This E. coli expresses LuxR constitutively and has GFP under Plux rep, thus it is supposed that GFP expression is repressed when AHL exists. Fig〇〇 shows that R0061 works as we expected.
(see more about Plux rep assay1...)
II, designing the new promoters repressed by LuxR/3OC6HSL, K395008 and K395009
Though R0061 is repressed by LuxR/3OC6HSL, the leaky expression is so high. That’s because -35 and -10 sequence of R0061 is the same as the -35 and -10 sequence of J23119 whose strength is the highest in BioBrick constitutive promoters.
Then we designed new BioBrick parts, K395008 and K395009, whose -35 and -10 sequence is different from R0061. The -35 and -10 sequence of K395008 is the same as that of J23108. The -35 and -10 sequence is the same as that of J23115. J23108 and J23115 are BioBrick constitutive promoters.
Why we chose J23108 and J23115 is that these expression levels are middle and low in BioBrick constitutive promoters. 2nd reason is that 3’ end nucleotide of -35 sequence of J23108 and J23115 is ‘A’ and that 5’ end nucleotide of -10 sequence 0f J23108 and J23115 is ‘T’. -35 and -10 overlap lux box.
III, characterization of K395008 (LuxR repression promoter)
Introduction
Even subtle changes in promoter may have distinct effects on the expression of gene. As we mentioned before, we designed a new promoter which is repressed by AHL and LuxR complex by changing one base of the existing BioBrick parts (BBa_R0061). We wanted to characterize this luxR repression promoter. Also, we wanted to confirm that this promoter is also repressed by AHL and LuxR but has different strength from the existing BioBrick part.
Result
After construction of K395195, we introduced K395195 and S03119 into DH5α. And we measured the fluorescence by flow cytometry. Fig 〇〇shows this result.
Addition of AHL caused the decrease in fluorescence intensity. The expression of GFP with AHL dropped to 1/3 comparing with the expression without AHL.
Conclusion
We confirmed that AHL repressed luxR repression promoter, K395008 as expected.
IV, characterization of R0062 (promoter activated by LuxR/3OC6HSL)
Introduction
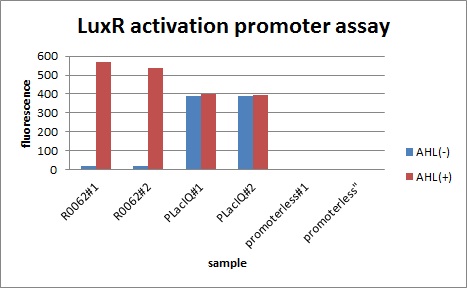
We characterized luxR activation promoter. We inserted chloramphenicol resistance coding sequence into the downstream of this promoter. This promoter plays an important role in Artificial Cooperation System. We wanted to confirm strength of this promoter which has already been done before in BioBrick in order to design new promoter based on this data. First, we assayed R0062 which is an existing BioBrick promoter activated by LuxR/3OC6HSL complex. To confirm this promoter, we constructed following two plasmids (fig〇〇and fig〇〇)
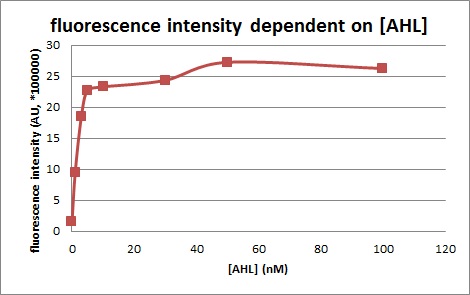
We introduced two plasmid into DH5α. Next, we measured the 3OC6HSL concentration dependence of Plux activity. We measured the fluorescence intensity under different concentration of AHL (0nM, 1nM, 3nM, 5nM, 10nM, 30nM, 50nM, 100nM. We measured the fluorescence by flow cytometry 3 hours after 3OC6HSL induction.
Result
In the absence of AHL, the fluorescence is low. In the contrast, the fluorescence is so high in the presence of 3OC6HSL(100nM), that’s 〇〇fold higher than in the absence of 3OC6HSL.
The previous experiment shows that Plux is worked. This means that Plux is activated by LuxR and 3OC6HSL.
Fig 〇〇is the result of measurement.
Conclusion
This E. coli expresses LuxR constitutively and has GFP under Plux act, thus it is supposed that GFP expression is activated when 3OC6HSL exists. We confirmed BioBricks, K395100 and S03119 (pSB3K3) worked correctly and the activity of R0062 is dependent on 3OC6HSL and the AHL threshold concentration is about 〇〇nM.
</div>
</div>
 "
"