Team:Tokyo Tech/Project/Artificial Cooperation System/luxI assay
From 2010.igem.org
3-3 luxI Assay
Contents |
Abstract
In order to test whether rbs-LuxI-term (K092400) works correctly, we measured the amount of 3OC6HSL synthesized by LuxI generator (K395146). We confirmed that rbs-LuxI-term (K092400) worked as expected and the amount of 3OC6HSL was sufficient to induce the chloramphenicol resistance gene expression at maximum level, which is regulated by R0062 in the Artificial Cooperation System.
Introduction
In the Artificial Cooperation System, where the amount of 3OC6HSL is an important parameter, enough amount of 3OC6HSL must be synthesized by a type of cell in order to induce the expression chloramphenicol resistance gene in the other type of cell.
Our aim in this experiment is to confirm that the amount of 3OC6HSL synthesized by the cell is sufficient to induce the expression chloramphenicol resistance gene in the Artificial Cooperation System. For this purpose, we constructed a constitutive LuxI generator PlacIq-LuxI (K395146) by ligating a constitutive promoter sequence PlacIq (I14032) to the upstream of rbs-LuxI-term (K092400) and measured the amount of 3OC6HSL synthesized with this device.
result
We confirmed that LuxI generator (K395146) caused the synthesis of 3OC6HSL and estimated the amount of 3OC6HSL by inducing GFP expressing reporter cell with the supernatant of culture of cell containing the LuxI generator. For this assay, we constructed a plasmid, expressing LuxI constitutively (K395146). In this plasmid PlacIq was chosen as a promoter due to following reason. Since we had already measured the expression activity ratio of PlacIq, R0062 (LuxR activation promoter) and K395008 (LuxR repression promoter), we can estimate the expression activity of the other promoters by measuring the expression activity of PlacIq.
After incubating the cell containing K395146 (PlacIq-LuxI), we collected the supernatant and divided it to induce 2 samples of the reporter cell. As a control, we cultured the reporter cell in two other different conditions; one was with induction of 3OC6HSL from reagent stock (positive control), and the other was without the induction of 3OC6HSL (negative control).
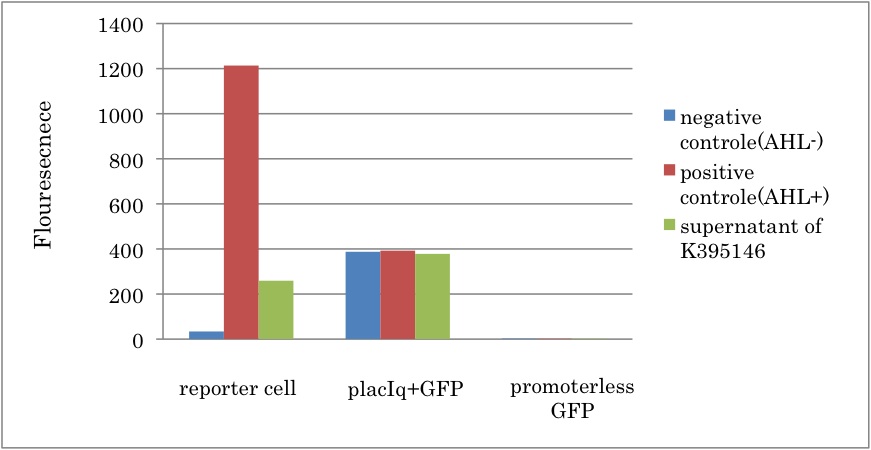
Fig.3-3-2 shows that GFP fluorescence increased in presence of the supernatant of liquid culture with LuxI-expression cell. This result proved that 3OC6HSL was synthesized by PlacIq-LuxI (K395146). In addition, we estimated the amount of 3OC6HSL produced by the cell with PlacIq-LuxI (K395146). By comparing Fig.3-3-2 with Fig3-1-2, it can be estimated that the concentration of 3OC6HSL synthesized by K395146 in each sample was almost 5nM. Since only half of the supernatant was used for induction in each sample., real concentration of synthesized 3OC6HSL was 10 nM.
Conclusion
From the experiment, we characterized the LuxI parts in the Registry. First, we corfirmed that rbs-LuxI-term (K092400) and LuxI generator (K395146) worked as expected. Second, we estimated that the concentration of 3OC6HSL in the supernatant of the cell containing K395146 was about 10nM, which is sufficient to induce the expression of chloramphenicol resistant gene.
Discussion
From comparison among each experiments, we estimated whether enough amount of AHL can be synthesized in Artificial cooperation system. According to results of this experiment and the concentration dependence of R0062 (promoter activated by LuxR/3OC6HSL), we found 10 nM AHL was synthesized. So, fig3-1-3 shows Lux repression promoter (R0061) is three-fold stronger than PlacIq in no repression. Replacements of LuxR-binding sequence of R0061 with LasR-binding one would turn it into Lux repression promoter which have similar strength as the Lux repression promoter. In that case, Plas(rep) – LuxI would be able to synthesize about 30 nM AHL.
This estimated AHL production from Plas(rep) – LuxI suggests that the Artificial cooperation system can synthesize the essential amount of AHL to rescue the counterpart cell in due to following two reasons. First, FigXX shows 30 nM AHL is sufficient to activate R0062 at maximum level. Thus 30 nM AHL can stimulate nearly same amount of chloramphenicol resistance protein as 100 nM AHL which is used in this XXX assay. Secondly, in the experiment of resistance gene activation device, the cell expressing chloramphenicol resistance gene was able to survive when initial concentration of AHL is 100nM. Taking into account that degradation rate of AHL is 0.6/h (ref), the concentration of AHL is less than 30 nM after 2~3 hours of start of the culture. Those estimations strongly suggest that AHL synthesized by E. coli probably induces an expression of chloramphenicol resistance gene and thus the growth of cell can recover. This is a most important point of the Artificial Cooperation System.
Materials&Methods
Construction
We replaced the pSB1A2 backborn of K092400 (RBS-LuxI-ter) with pSB6A1, and ligated I14032 (PlacIq) to the upstream of K092400. Then we introduced the constructed plasmid, K395146, into DH5α.
Samples
- [Plux act - GFP](BBa_K395100) on pSB6A1 + [Ptet – LuxR] on pSB3K3 = reporter cell
- [PlacIq-luxI](BBa_K395146) on pSB6A1 = inducer cell
Strain
DH5 DH5α.
Protocol
- Prepare overnight culture at 37℃ for 12hours. (2 tubes for each sample)
- Take 30 μl of the overnight culture of reporter cell into LB + antibiotics (Amp + Kan). (→fresh culture)
- Incubate the flesh culture of reporter cell] until the observed O.D. reaches around 0.60. and gather the supernatant of culture of inducer cell.
- Each sample of reporter cell was divided into 2. Prepare and add AHL mixture to one, cell-synthesized AHL to another one, and DMSO mixture to the other.
- Induction of reporter cell for 3 hours.
- Fluorometer (FLA5200) and flow cytometry measurements for GFP expression of reporter cell.
Reference
- Ying-jin Yuan et al PLoS 2010, e10619
- W. Shaw, et al Br. Med. Bull. 1984, 40, 36.
 "
"