Team:Tokyo Tech/Project/Artificial Cooperation System
From 2010.igem.org
(→Abstract) |
|||
| Line 45: | Line 45: | ||
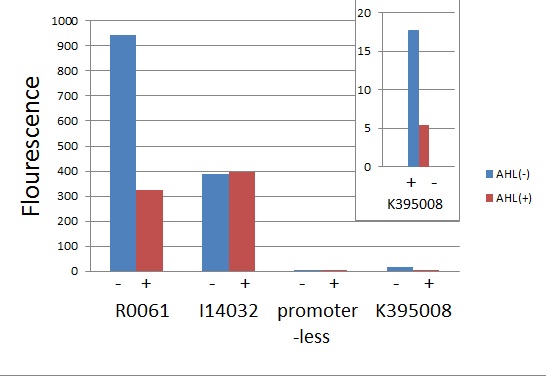
[[IMAGE:Tokyotech_R0061_K395008_graph.jpg|265px|left|thumb|Fig1 LuxR Repression promoter assay. This work done by Eriko Uchikoshi and Shohei Kitano]] | [[IMAGE:Tokyotech_R0061_K395008_graph.jpg|265px|left|thumb|Fig1 LuxR Repression promoter assay. This work done by Eriko Uchikoshi and Shohei Kitano]] | ||
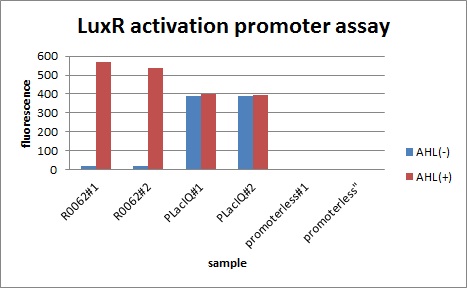
[[IMAGE:tokyotech_LuxR activation promoter assay(R0062).jpg|300px|left|thumb|Fig2 LuxR activation promoter assay. This work done by Eriko Uchikoshi and Shohei Kitano]] | [[IMAGE:tokyotech_LuxR activation promoter assay(R0062).jpg|300px|left|thumb|Fig2 LuxR activation promoter assay. This work done by Eriko Uchikoshi and Shohei Kitano]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
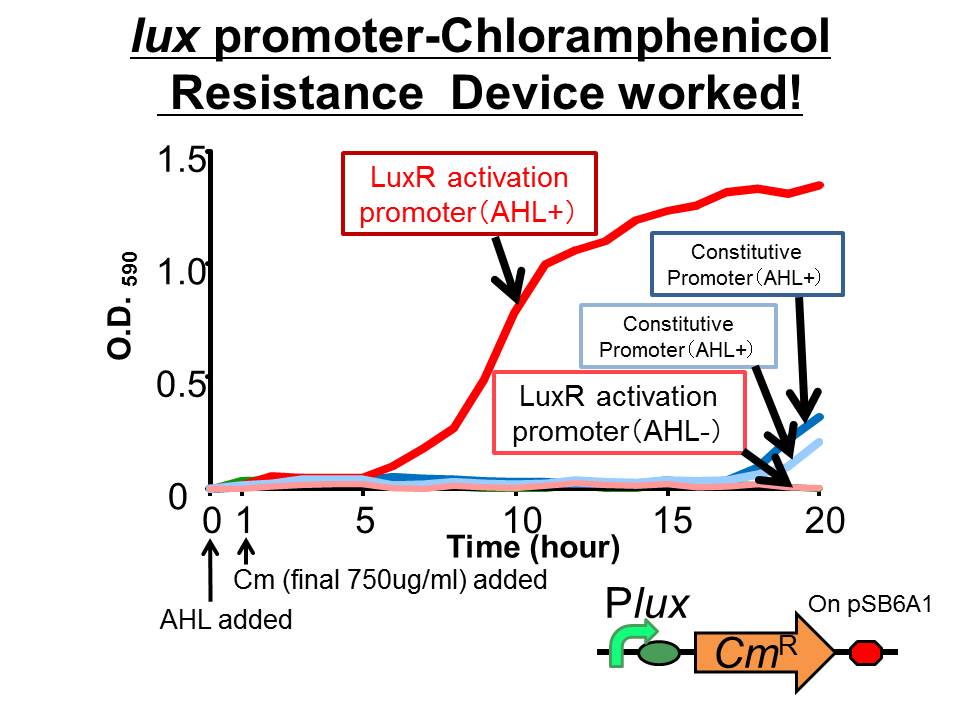
| + | [[Image:tokyotech_Cm-survival.jpg|265px|left|thumb|Fig3 Chloramphenicol resistance device. This work done by Yusuke Kaneta]] | ||
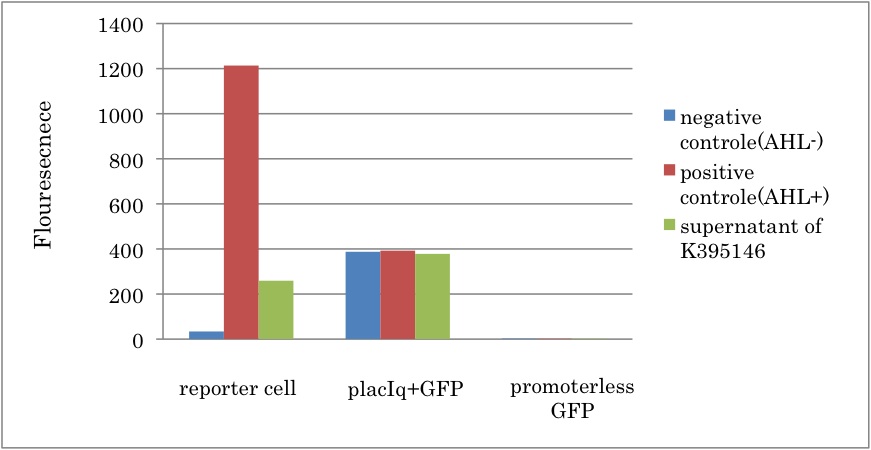
| + | [[IMAGE:Tokyotech Fig.K395146-1.jpg|300px|left|thumb|Fig4 LuxI and LuxR activation promoter assay. This work done by Eriko Uchikoshi and Shohei Kitano]] | ||
=Introduction= | =Introduction= | ||
Revision as of 15:28, 27 October 2010
3 Artificial Cooperation System
Contents |
Artificial Cooperation System
Abstract
We built Artificial Cooperation System to engineered E.coli with humanity. In this system quorum sensing and chloramphenicol generator activated by LuxR are very important. First, we constructed and characterized several promoters regulated by LuxR and 3OC6HSL. We designed and constructed a new LuxR repression promoter. Also, we characterized the functions of a LuxR repression promoter and a LuxR activation promoter in the registry. Second, we constructed a new chloramphenicol resistance gene generator which is regulated by LuxR activation promoter. We confirmed that the cell with the chloramphenicol resistance gene generator under high concentration of chloramphenicol when 3OC6HSL exists. Third, we confirmed the feasibility of Artificial Cooperation System from simulation and experimental data.
Introduction
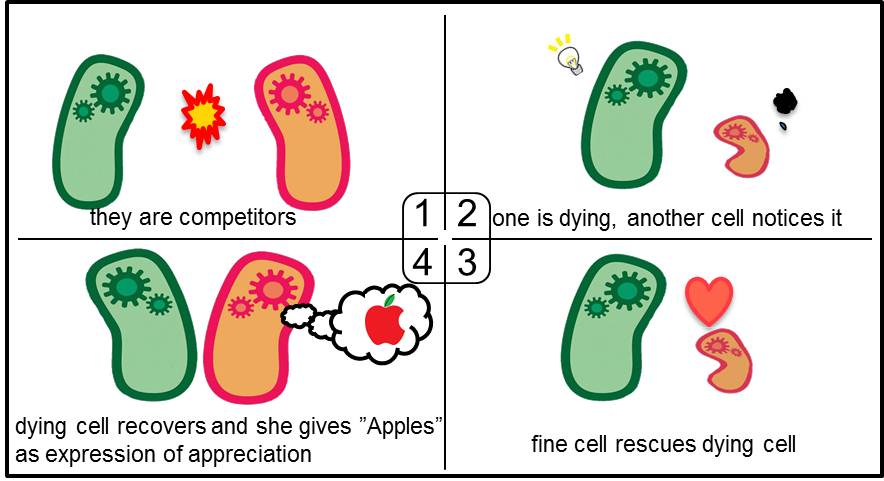
What’s Artificial Cooperation System?
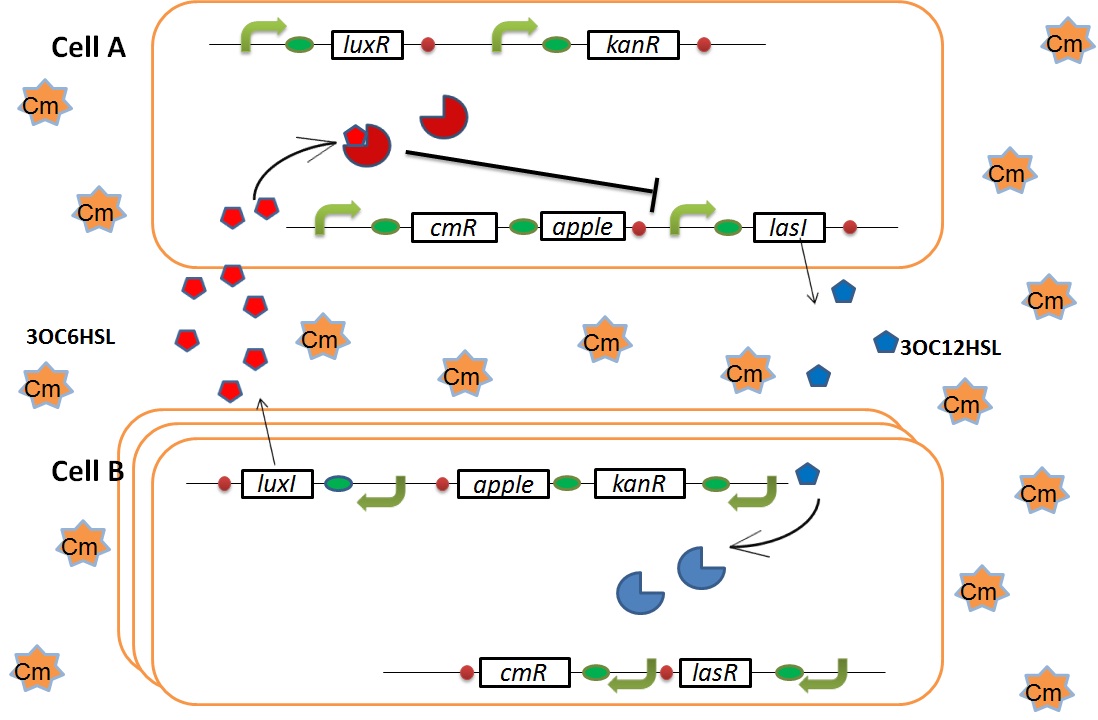
Artificial Cooperation System was built to engineer E. coli that behaves kindly and have sympathy. This system consists of two types of cells. In normal conditions, they are competitors for survival. While in critical conditions, such that one is dying, another cell would notice the crisis of dying cell and help her. This is a summary of the Artificial Cooperation System.
Genetic Circuit
I, genetic circuit overview
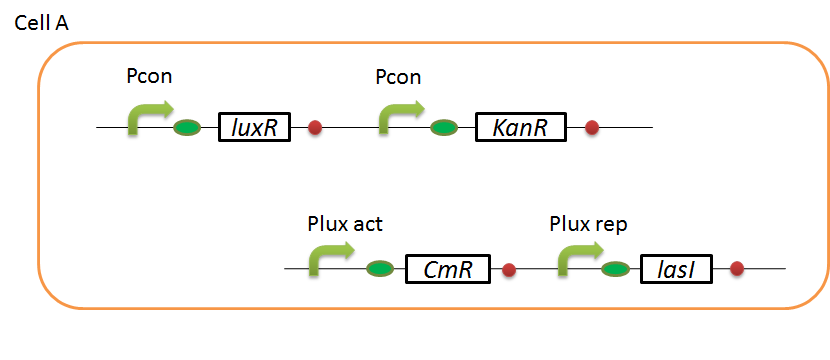
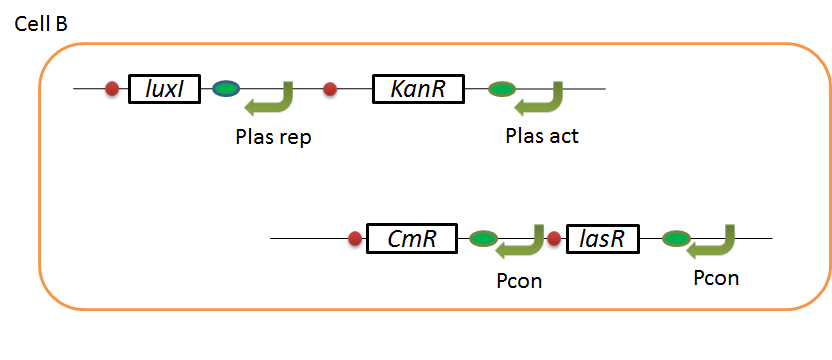
We designed the following circuit. In Artificial Cooperation System, there are two types of E. coli, Cell A and Cell B. Cell A has resistance to kanamycin and Cell B has resistance to chloramphenicol originally. (See more…)
Pcon: constitutive promoter.
Plux act: promoter activated by LuxR and 3OC6HSL. We call this promoter luxR activation promoter.
Plux rep: promoter repressed by LuxR and 3OC6HSL. We call this promoter luxR repression promoter.
Plas act: promoter activated by LasR and 3OC12HSL. We call this promoter lasR activation promoter.
Plas rep: promoter repressed by LasR and 3OC12HSL. We call this promoter lasR repression promoter.
lasI: enzyme repressed by LuxR and 3OC6HSL. We call this promoter LuxR repression promoter.
luxI: enzyme repressed by LasR and 3OC12HSL. We call this promoter LasR repression promoter.
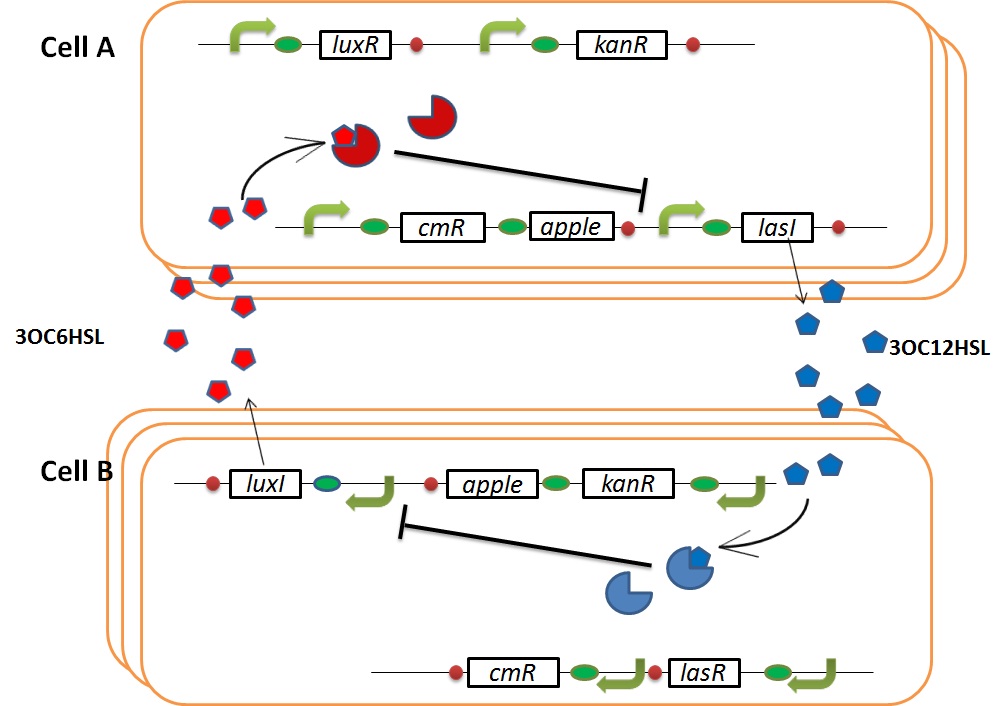
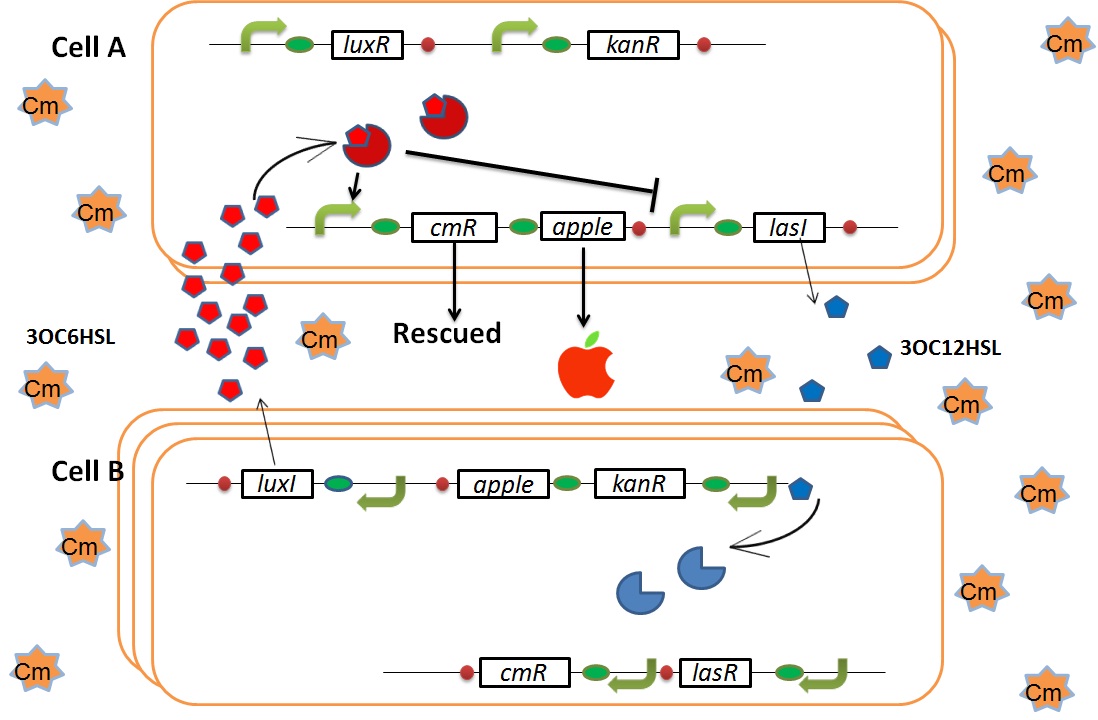
In cell A, production of LasI is repressed by LuxR/3OC6HSL complex and that of Cm resistance gene is activated by LuxR/3OC6HSL complex. In cell B, production of LuxI is repressed by LasR/3OC12HSL complex and that of Kan resistance gene is activated by LasR/3OC12HSL complex. This means that 3OC12HSL produced by cell A and 3OC6HSL produced by cell B indirectly repress each other’s production. And 3OC12HSL activates resistance gene of cell B while 3OC6HSL activates resistance gene of cell A. Apple gene is composed of crtEBIYZW and MpAAT1. This expresses only when the cell is rescued and recovered.
Normal
In normal situation, Cell A and Cell B are competitors and recognize each other by using quorum sensing. First, LasI protein in Cell A produces 3OC12HSL. Second, these signal molecules diffuse through cell membranes and bind to LasR protein in Cell B. Third, LasR/3OC12HSL complex represses the production of LuxI protein. Similarly in cell B, LuxI protein produces 3OC6HSL. These signal molecules diffuse through cell membranes and bind to LuxR protein in Cell A. LuxR/3OC6HSL complex represses the production of LasI protein. Although Cell A and Cell B have chloramphenicol and kanamycin resistance gene respectively, they don’t express in this situation. This is because the concentration of 3OC12HSL and 3OC6HSL is too low to activate the expressions of kanamycin resistance gene and chloramphenicol resistance gene, respectively.
when Cell A is dying
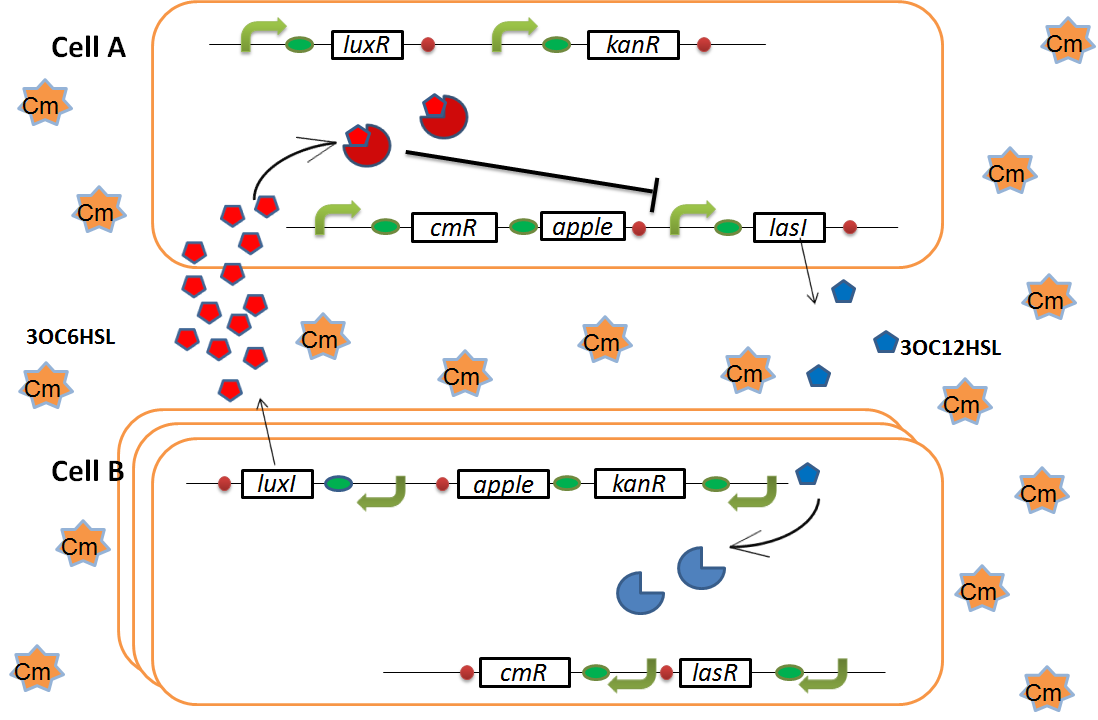
As we mentioned, originally Cell A has a resistance gene against kanamycin and does not have that against chloramphenicol. On the other hand, Cell B has resistance to chloramphenicol and doesn’t have resistance to kanamycin. Therefore, the addition of chloramphenicol only decreases the number of Cell A. It means that 3OC12HSL decreases. It also results in the decrease of LasR/3OC12HSL complex that repress the expression of LasI in Cell B. By this process, Cell B notices that Cell A is dying and tries to rescue it. when Cell B tries to rescue Cell A
As we mentioned, originally Cell A has a resistance gene against kanamycin and does not have that against chloramphenicol. On the other hand, Cell B has resistance to chloramphenicol and doesn’t have resistance to kanamycin. Therefore, the addition of chloramphenicol only decreases the number of Cell A. It means that 3OC12HSL decreases. It also results in the decrease of LasR/3OC12HSL complex that repress the expression of LasI in Cell B. By this process, Cell B notices that Cell A is dying and tries to rescue it.
plux-CmR
We succeeded in construction and characterization of a NEW Biobrick device of a chloramphenicol resistance (CmR) generator ([http://partsregistry.org/Part:BBa_K395162 BBa_K395162]) which is activated by LuxR activation promoter. Fig〇〇 shows that in the presence of 3OC6HSL, the device is activated and as a result the cell was able to survive under high chloramphenicol concentration (750 ug/ml). See more
luxI
We constructed a new LuxI generator (K395146) and characterized its function. By combining I14032 (PlacIq) and K092400 (rbs-luxI-ter), we constructed K395146. Then we confirmed that the cell with K395146 produced the sufficient amount of 3OC6HSL to activate LuxR activation promoter (R0062) See more
Mathematical modeling
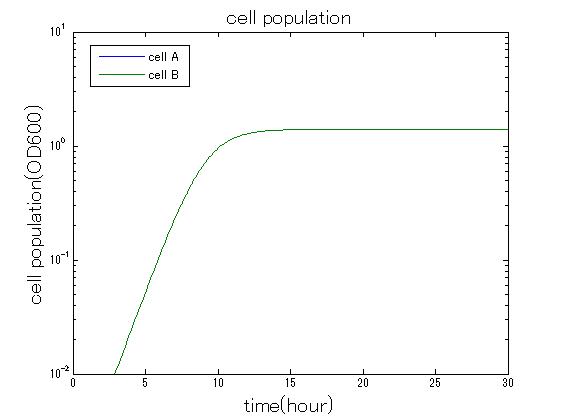
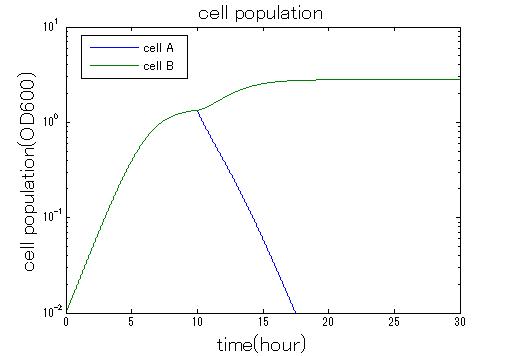
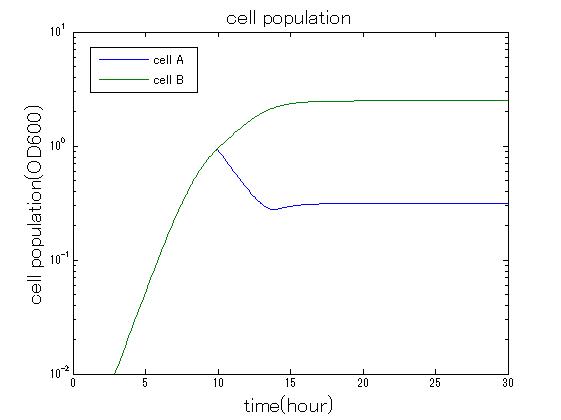
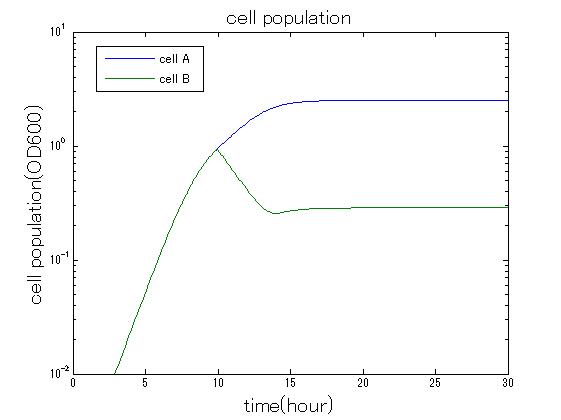
In order to confirm that this system is feasible, we simulated this system. The following figures 〇〇 show the simulation result. We held a series of simulations under several experimental conditions. Fig◯◯〜Fig◯◯ are the results. Fig1 is the case without antibiotic. In this case, cell A and cell B show no difference of growth. Fig2 is the condition with chloramphenicol while Artificial Cooperation System is not working. Graph shows that cellA decrease by chloramphenicol. Fig.3 is the condition with chloramphenicol while Artificial Cooperation System is working. The graph shows turnover of CellA, which means it was rescued by Artificial Cooperation System. Fig4 is the condition with Kanamycin while Artificial Cooperation System is working. This time, number of CellB decrease at first, though recover at the end. From the simulation results above, we can conclude that our system can work under a certain situation.
 "
"