Team:Tokyo Tech/Project/Apple Reporter2
From 2010.igem.org
(→Materials and Methods) |
|||
| Line 90: | Line 90: | ||
=Materials and Methods= | =Materials and Methods= | ||
| - | Strains of | + | Strains of ''E. coli''<Br>''E. coli'' BL21 DE3 |
| - | + | ||
| - | Varieties of plasmid | + | Varieties of plasmid<Br> |
| - | MpAAT1 on pSB6A1 | + | MpAAT1 on pSB6A1<Br> |
Trx on pACYC184 | Trx on pACYC184 | ||
| - | Substrate | + | Substrate<Br> |
| - | Butanol (final 0.4%) | + | Butanol (final 0.4%)<Br> |
| - | 2-methyl butanol (final 0.2%) | + | 2-methyl butanol (final 0.2%)<Br> |
| - | Inducer | + | Inducer<Br> |
| - | 100 mM IPTG | + | 100 mM IPTG<Br> |
| - | 20% arabinose | + | 20% arabinose<Br> |
| - | Solution | + | Solution<Br> |
| - | Undecane solution: undecane 10 | + | Undecane solution: undecane 10 μL + ether 990 μL |
| - | MpAAT1 expression construct. | + | MpAAT1 expression construct.<Br> |
| - | We ordered the synthesis of ‘’MpAAT1’’([http://partsregistry.org/Part:BBa_K395602 BBa_K395602]) from Mr. GENE. | + | We ordered the synthesis of ‘’MpAAT1’’([http://partsregistry.org/Part:BBa_K395602 BBa_K395602]) from Mr. GENE.<Br> |
| - | This artificial gene was annealed with vector pSB6A1 as MpAAT1 expression plasmid. | + | This artificial gene was annealed with vector pSB6A1 as MpAAT1 expression plasmid.<Br> |
| - | Moreover, we introduced pTrx6 into this expression plasmid to stabilize the ‘’MpAAT1’’ gene product. | + | Moreover, we introduced pTrx6 into this expression plasmid to stabilize the ‘’MpAAT1’’ gene product.<Br> |
| - | MpAAT1 over expression conditions | + | MpAAT1 over expression conditions<Br> |
| - | Artificial gene has T7 promoter on the upstream of ‘’MpAAT1’’. | + | Artificial gene has T7 promoter on the upstream of ‘’MpAAT1’’.<Br> |
| - | This promoter works by taking over T7 RNA polymerase from | + | This promoter works by taking over T7 RNA polymerase from ''E. coli''.<Br> |
| - | Therefore we utilized | + | Therefore we utilized ''E. coli'' BL21 DE3 which has T7 RNA polymerase.<Br> |
| - | Furthermore, arabinose was added in culture to induce Trx which has arabinose-induced promoter. | + | Furthermore, arabinose was added in culture to induce Trx which has arabinose-induced promoter.<Br> |
| - | + | ''E. coli'' BL21 DE3 <Br> | |
| - | + | ''E. coli'' BL21 DE3 express T7 RNA polymerase by IPTG induction.<Br> | |
| - | Therefore we added 3 | + | Therefore we added 3 μL of 100 mM IPTG and 15 μL of 20% arabinose in LB culture in order to express MpAAT1 and Trx in ''E. coli'' BL21 DE3.<Br> |
| - | Expression of MpAAT1 recombinant protein in | + | Expression of MpAAT1 recombinant protein in ''E. coli''<Br> |
| - | O/N | + | O/N ''E. coli''<Br> |
| - | →100-fold dilution | + | →100-fold dilution <Br> |
| - | →2 hour fresh culture | + | →2 hour fresh culture<Br> |
| - | →make up the OD<sub>590</sub> = 0.1 with LB culture | + | →make up the OD<sub>590</sub> = 0.1 with LB culture<Br> |
| - | →add substrate,antibiotics and inducer | + | →add substrate,antibiotics and inducer<Br> |
| - | →O/N | + | →O/N<Br> |
| - | →harvest (7,000×’’g’’, 3 min) | + | →harvest (7,000×’’g’’, 3 min)<Br> |
| - | →collect supernatant solution | + | →collect supernatant solution<Br> |
| - | →separating liquid layer and oil layer with ether (shake supernatant solution 0.5 mL, ether 0.5 mL and undecane soln. 2 | + | →separating liquid layer and oil layer with ether (shake supernatant solution 0.5 mL, ether 0.5 mL and undecane soln. 2 μL)<Br> |
| - | →derive oil layer from supernatant solution with liquid-liquid extraction | + | →derive oil layer from supernatant solution with liquid-liquid extraction<Br> |
| - | →GC analysis. | + | →GC analysis.<Br> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | GC analysis<Br> | ||
| + | GC: SHIMADZU GAS CHROMATOGRAPH GC-14B<Br> | ||
| + | Column: J&W SCIENTIFIC, DB-17, Film thickness 0.25 μm, Column Dimensions 15 m * 0.320 mm, Temperature Limits 40°C to 280oC (300oC Program)<Br> | ||
| + | Conditions: column temperature 35°C, injector temperature 180°C, detector temperature 180°C<Br> | ||
| + | Sample was injected 5 μL.<Br> | ||
=reference= | =reference= | ||
Revision as of 16:29, 27 October 2010
2-2 Fragrance
Contents |
Abstract
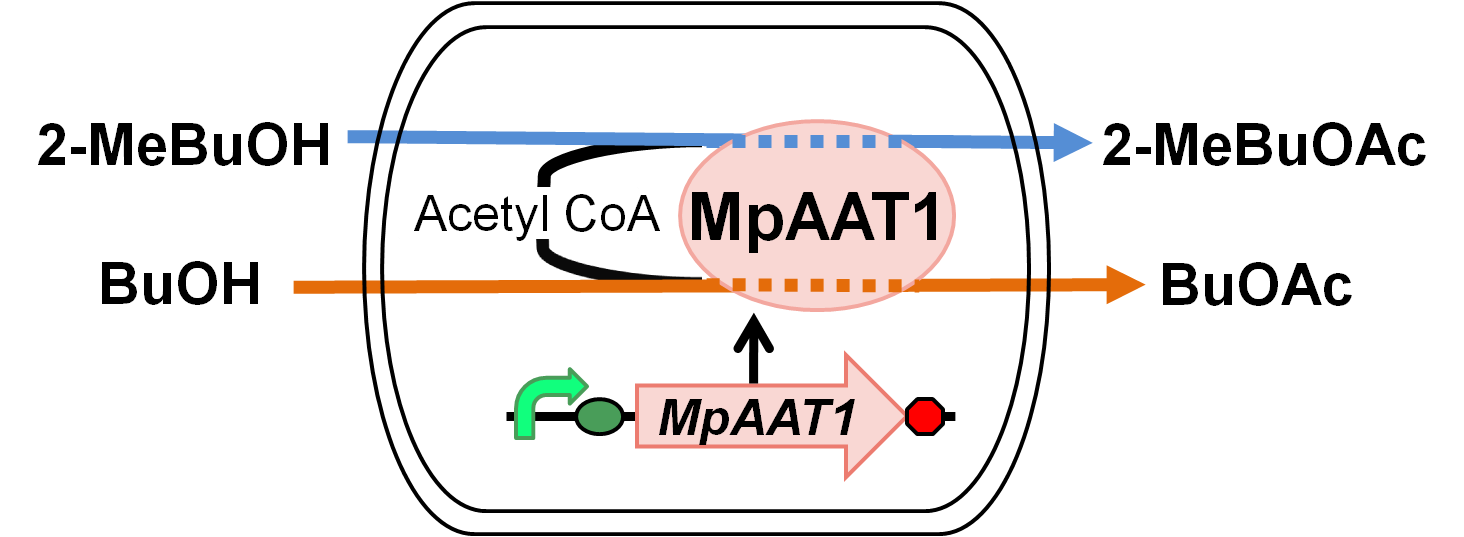
We designed apple fragrance expression device with MpAAT1[1]. Fig. 2-2-1 shows the outline of the device. MpAAT1 converts substrate and Acetyl CoA into apple fragrance molecules. Acetyl CoA exists originally in ‘’E.coli’’ cell. MpAAT1 converts 2-methyl butanol and butanol into 2-methylbutyl acetate and butyl acetate respectively.
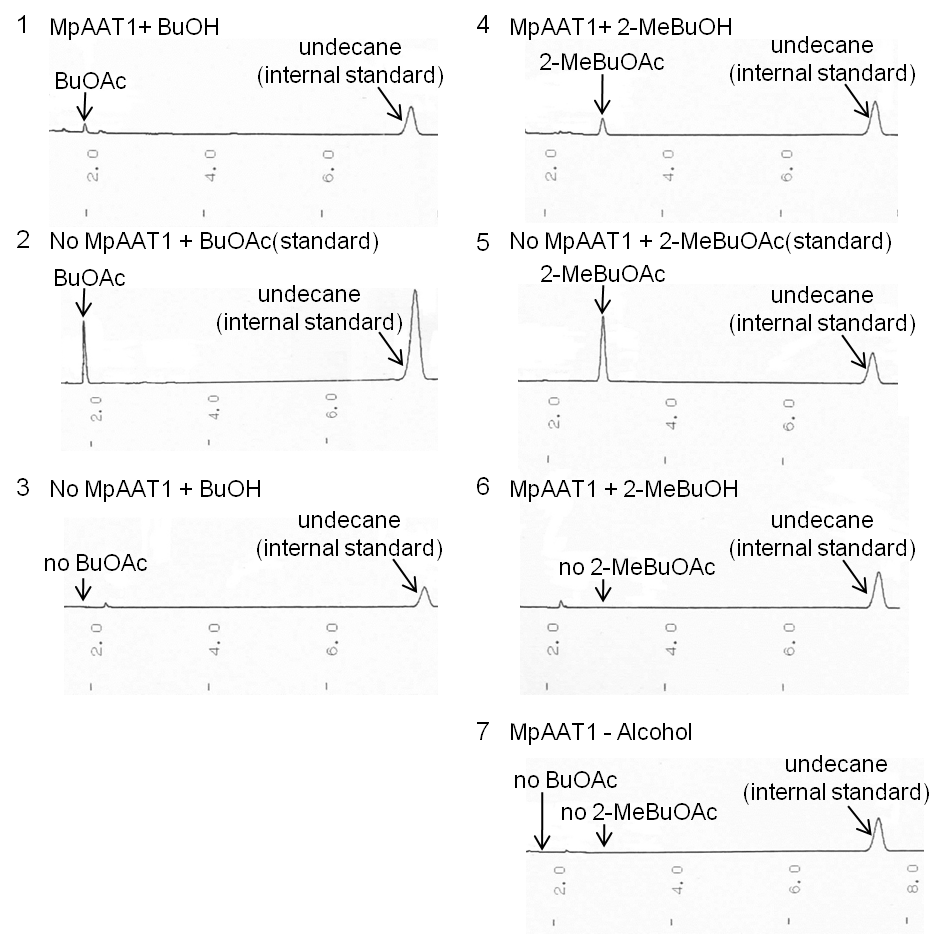
We performed gas chromatography to confirm the production of the esters, and the results revealed that MpAAT1 successfully converted alcohol and Acetyl CoA into our target esters (Fig. 2-2-2).
Introduction
‘’MpAAT1’’ gene was isolated from ‘’Malus pumila’’ (cv. Royal Gala). This gene is expressed in leaves, flowers and fruit of apple. The recombinant enzyme (MpAAT1) utilize various alcohol as substrates such as straight chain (C3–C10), branched chain alcohols, aromatic alcohols and terpene alcohols. For producing esters, various kinds of CoA are used, either. Both alcohol and CoA are required for successful ester synthesis. The story is, when dying cells are recued by its counterpart, they give an apple for a token of their gratitude. The major components for producing apple fragrance are butyl acetate and 2-methylbutyl acetate. In this project, we engineered ‘’E.coli’’ to produce these molecules by MpAAT1.
Result
We transformed MpAAT1([http://partsregistry.org/Part:BBa_K395602 BBa_K395602]) on pSB6 along with pTrx6 into ‘’E.coli’’ BL21 DE3, and cultured after addition of alcohols(2-MeBuOH or BuOH) as substrates.
After 12 hours of incubation, we extracted organic soution layer from the culture and analyzed by gas chromatography (Fig. 2-2-2). Peaks of the esters producing apple fragrance (2-MeBuOAc or BuOAc) were detected.
(A) In (2), the peak of BuOAc is shown. Comparing with (1), the position of the peak is in the same position. From this result, we can see MpAAT1 converted BuOH into BuOAc. (B) In (3), we observed the peak of BuOH, which we are going to use as substrate.This graph doesn’t show peak in the same place with that of BuOAc in (1).This means BuOH used as substrate doesn’t contain BuOAc. (C) In the same way as (A), from comparison of (4) and (5), we can see 2-MeBuOAC is synthesized from 2-MeBuOH. (D) (6) shows the peak of 2-MeBuOH, though no peak of 2-MeBuOAc. Thus, 2-MeBuOH doesn’t contain 2- MeBuOAc. (E) In (7), no peak of ester is observed. This sample wasn’t added with substrates, so MpAAT1 without substrates didn’t produce esters. From these results, we can conclude that E.Coli with MpAAT1 is able to convert substrate alcohols into esters. ↑ どっちがいいですか ↓ (A)In (2), the peak of BuOAc is shown. Comparing with (1), the position of the peak is in the same position. From this result, we can see MpAAT1 converted BuOH into BuOAc. (B)In (3), we observed the peak of BuOH, which we are going to use as substrate. This graph doesn’t show peak in the same place with that of BuOAc in (1). This means BuOH used as substrate doesn’t contain BuOAc . (C)In the same way as 1., from comparison of (4)and (5), we can see 2-MeBuOAC is synthesized from 2-MeBuOH. (D)(6) shows the peak of 2-MeBuOH, though no peak of 2-MeBuOAc.Thus, 2-MeBuOH doesn’t contain2- MeBuOAc. (E)(7), no peak of ester is observed. This sample wasn’t added with substrate, so MpAAT1 without substrate didn’t produce esters. From these results, we can conclude that E.Coli with MpAAT1 is able to convert substrate alcohol into esters.
Discussion
From the result of the experiment above, we can conclude that engineered ‘’E.coli’’ successfully converted alcohols added as substrates into esters. Moreover, we synthesized MpAAT1 and esters using ‘’E.coli’’ BL21 DE3 as a chassis, which was reported to be implausible in former report(s)[1]. In 2008, C.R. Shen and J.C. Liao[2] succeeded in synthesizing butanol from ‘’E.coli’’. If we take advantage of this engineered ‘’E.coli’’, we could produce apple fragrance ester without the addition of substrate.
Materials and Methods
Strains of E. coli
E. coli BL21 DE3
Varieties of plasmid
MpAAT1 on pSB6A1
Trx on pACYC184
Substrate
Butanol (final 0.4%)
2-methyl butanol (final 0.2%)
Inducer
100 mM IPTG
20% arabinose
Solution
Undecane solution: undecane 10 μL + ether 990 μL
MpAAT1 expression construct.
We ordered the synthesis of ‘’MpAAT1’’([http://partsregistry.org/Part:BBa_K395602 BBa_K395602]) from Mr. GENE.
This artificial gene was annealed with vector pSB6A1 as MpAAT1 expression plasmid.
Moreover, we introduced pTrx6 into this expression plasmid to stabilize the ‘’MpAAT1’’ gene product.
MpAAT1 over expression conditions
Artificial gene has T7 promoter on the upstream of ‘’MpAAT1’’.
This promoter works by taking over T7 RNA polymerase from E. coli.
Therefore we utilized E. coli BL21 DE3 which has T7 RNA polymerase.
Furthermore, arabinose was added in culture to induce Trx which has arabinose-induced promoter.
E. coli BL21 DE3
E. coli BL21 DE3 express T7 RNA polymerase by IPTG induction.
Therefore we added 3 μL of 100 mM IPTG and 15 μL of 20% arabinose in LB culture in order to express MpAAT1 and Trx in E. coli BL21 DE3.
Expression of MpAAT1 recombinant protein in E. coli
O/N E. coli
→100-fold dilution
→2 hour fresh culture
→make up the OD590 = 0.1 with LB culture
→add substrate,antibiotics and inducer
→O/N
→harvest (7,000×’’g’’, 3 min)
→collect supernatant solution
→separating liquid layer and oil layer with ether (shake supernatant solution 0.5 mL, ether 0.5 mL and undecane soln. 2 μL)
→derive oil layer from supernatant solution with liquid-liquid extraction
→GC analysis.
GC analysis
GC: SHIMADZU GAS CHROMATOGRAPH GC-14B
Column: J&W SCIENTIFIC, DB-17, Film thickness 0.25 μm, Column Dimensions 15 m * 0.320 mm, Temperature Limits 40°C to 280oC (300oC Program)
Conditions: column temperature 35°C, injector temperature 180°C, detector temperature 180°C
Sample was injected 5 μL.
reference
[1] Edwige J. F. Souleyre, FEBS Journal, 272, 3132–3144 (2005) [2] Shota Atsumi, Metabolic Engineering, 10, 305–311 (2008) [3] Young H, J Sci Food Agric, 71, 329-336 (1996)
 "
"