Team:Paris Liliane Bettencourt/Project/Population counter/Results
From 2010.igem.org
(Difference between revisions)
| Line 3: | Line 3: | ||
<html> | <html> | ||
| - | |||
<p style="display:block;"> | <p style="display:block;"> | ||
<a href="https://2010.igem.org/Team:Paris_Liliane_Bettencourt/Project/Population_counter"> | <a href="https://2010.igem.org/Team:Paris_Liliane_Bettencourt/Project/Population_counter"> | ||
| - | <img src="https://static.igem.org/mediawiki/2010/ | + | <img src="https://static.igem.org/mediawiki/2010/3/30/Popcount.png" width="75" height="75" title="Population Counter"> |
</a> | </a> | ||
| - | <font size=4>Population counter</font> | + | <font size=4>Population counter : Results</font> |
<a href="https://2010.igem.org/Team:Paris_Liliane_Bettencourt/Project/SIP"> | <a href="https://2010.igem.org/Team:Paris_Liliane_Bettencourt/Project/SIP"> | ||
| - | <img src="https://static.igem.org/mediawiki/2010/4/4c/SIP.png" width=" | + | <img src="https://static.igem.org/mediawiki/2010/4/4c/SIP.png" width="75" height="75" align=right title="SIP"> |
</a> | </a> | ||
<a href="https://2010.igem.org/Team:Paris_Liliane_Bettencourt/Project/Memo-cell"> | <a href="https://2010.igem.org/Team:Paris_Liliane_Bettencourt/Project/Memo-cell"> | ||
| - | <img src="https://static.igem.org/mediawiki/2010/ | + | <img src="https://static.igem.org/mediawiki/2010/e/e8/Memocell.png" width="75" height="75" align=right title="Memo-Cell"> |
</a> | </a> | ||
<br /> | <br /> | ||
| Line 45: | Line 44: | ||
<br><br></html> | <br><br></html> | ||
*Check if IntI1 integrase works | *Check if IntI1 integrase works | ||
| - | + | <br> Then we wanted to see if the IntI1 integrase actually works. We have done a double transformation "RFP Counter" (on pSB1A2 which is a high copy plasmid) + "pBAD-IntI1 (which is pSulib on a low copy plasmid With Kanamycin resistance) on a plate containing ampicillin, kanamycin and 1% glucose (which blocks the pBAD promoter and avoid recombination events to occur before the test of our counter). The protocol is quite simple : we dilute an overnight culture (without glucose otherwise pBad cannot be activated), wait until OD 0,2 and add arabinose to begin the test (t=0h). At regular time intervals, we plated on plates containing ampicillin and 1% glucose (this is not important if the cells lose the "pBad-Int" plasmid because we don't want recombinations to happen on the plates). | |
| - | <br>[[Image:ara1_1.jpg|150px|]] [[Image:ara1_2.jpg|150px|]] [[Image:ara1_3.jpg|150px|]] [[Image:ara1_4.jpg|150px|]] | + | <br> |
| - | < | + | <tr> |
| - | <br>[[Image:ara1_5.jpg|150px|]] [[Image:ara1_6.jpg|150px|]] [[Image:ara1_7.jpg|150px|]] [[Image:ara1_8.jpg|150px|]] | + | <td>[[Image:ara1_1.jpg|150px|]]</td> <td>[[Image:ara1_2.jpg|150px|]]</td> <td>[[Image:ara1_3.jpg|150px|]]</td> <td>[[Image:ara1_4.jpg|150px|]]</td> |
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1h</td> <td>2h</td> <td>3h</td> <td>4h</td> | ||
| + | <tr> | ||
| + | <br><br> | ||
| + | [[Image:ara1_5.jpg|150px|]] [[Image:ara1_6.jpg|150px|]] [[Image:ara1_7.jpg|150px|]] [[Image:ara1_8.jpg|150px|]] | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>5h</td> <td>6h</td> <td>7h</td> <td>8h</td> | ||
| + | </tr> | ||
| + | <br> | ||
| + | </tr> | ||
| + | |||
<br><br> | <br><br> | ||
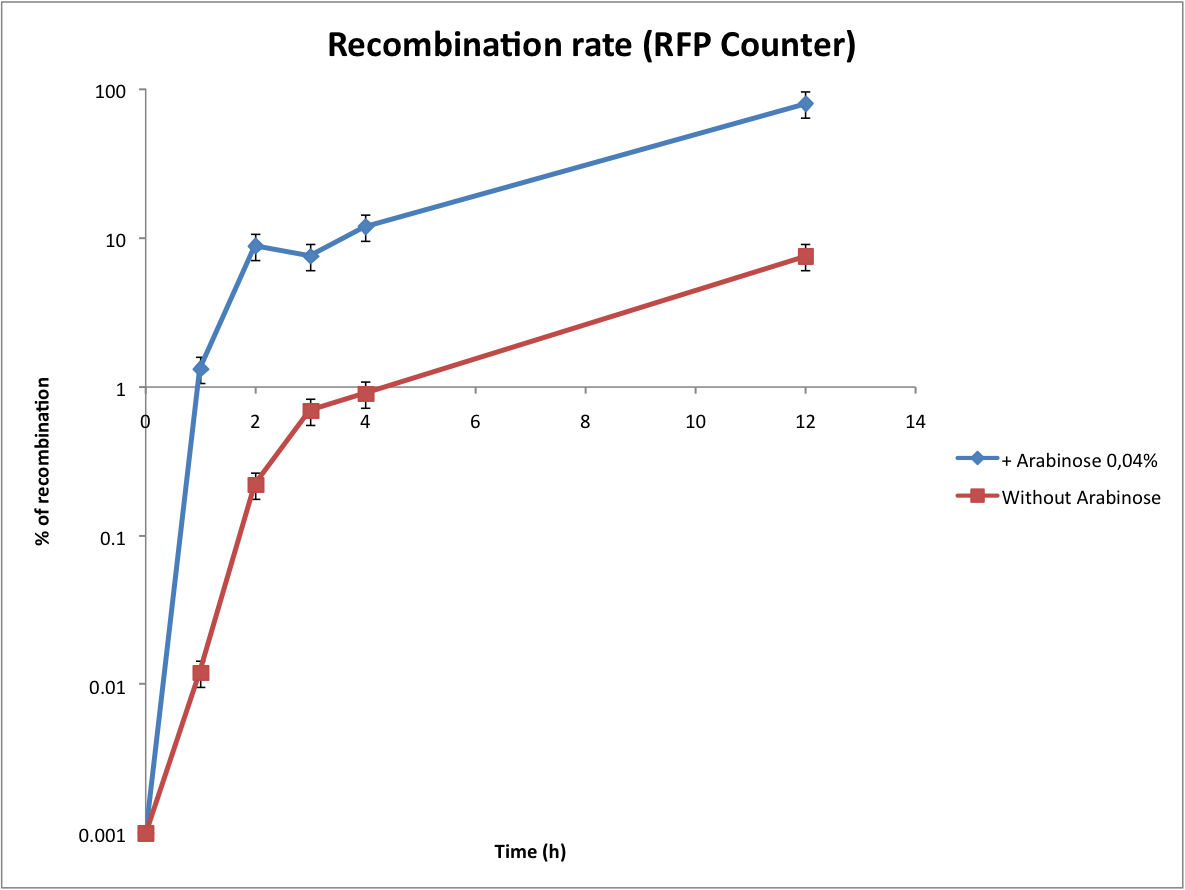
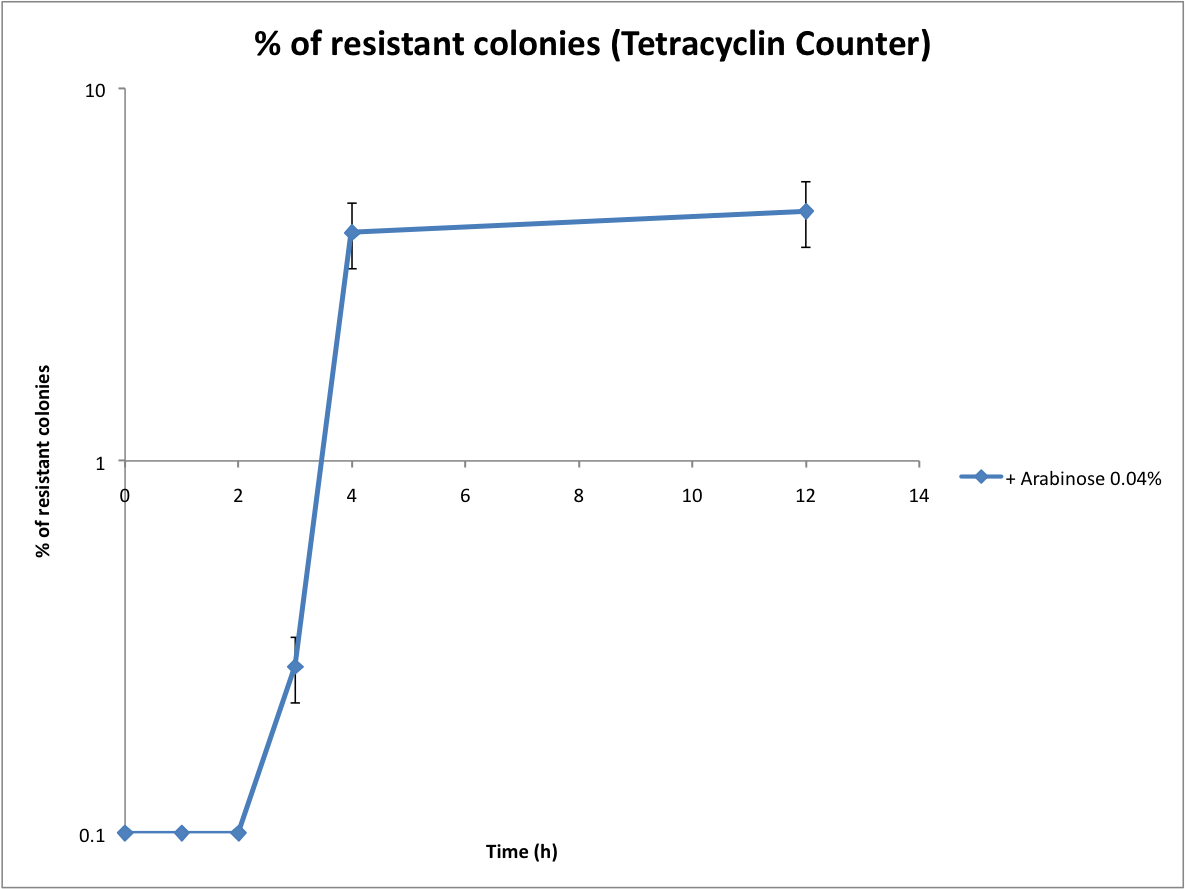
*Calculate the recombination rate | *Calculate the recombination rate | ||
Revision as of 20:21, 27 October 2010
- Construction of our counters
First of all, we spent a significant part of the summer to do cloning. For this, we have used a lot of biobricks submitted to the parts registry by previous teams, thank you! Using these parts we have created plenty of new composite parts that are highly modular (different kind of RBS and promoters) and could serve next iGEM teams. We have also created some new biobricks that are at the core of our project. We hope other iGEM teams will work with integrons and might need these parts.
Moreover, each of our constructs was verified by sequencing. For more information you can take a look at the parts that we sent to the registry.
- Check if IntI1 integrase works
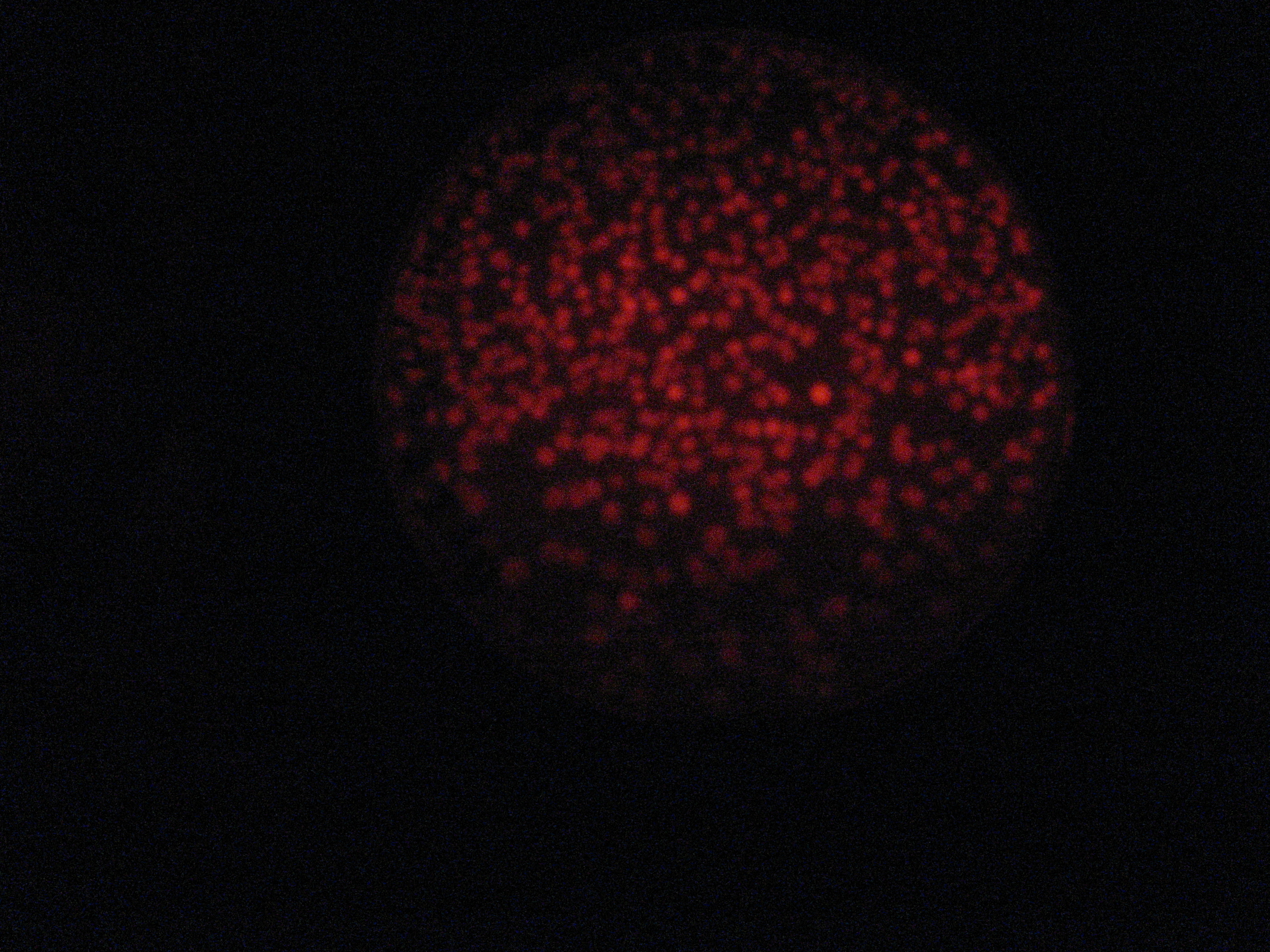
Then we wanted to see if the IntI1 integrase actually works. We have done a double transformation "RFP Counter" (on pSB1A2 which is a high copy plasmid) + "pBAD-IntI1 (which is pSulib on a low copy plasmid With Kanamycin resistance) on a plate containing ampicillin, kanamycin and 1% glucose (which blocks the pBAD promoter and avoid recombination events to occur before the test of our counter). The protocol is quite simple : we dilute an overnight culture (without glucose otherwise pBad cannot be activated), wait until OD 0,2 and add arabinose to begin the test (t=0h). At regular time intervals, we plated on plates containing ampicillin and 1% glucose (this is not important if the cells lose the "pBad-Int" plasmid because we don't want recombinations to happen on the plates).
<tr> <td>
 </td> <td>
</td> <td>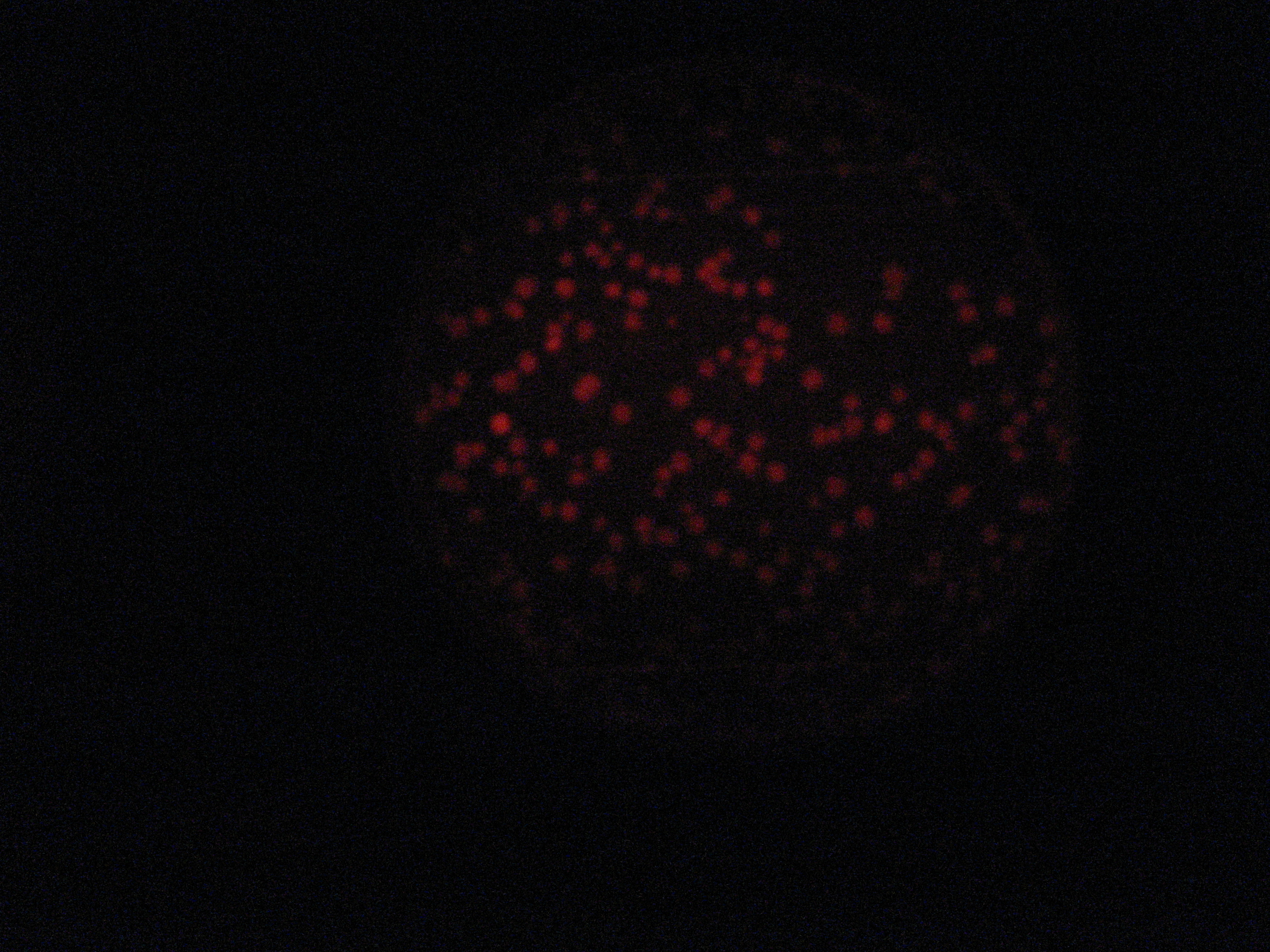 </td> <td>
</td> <td>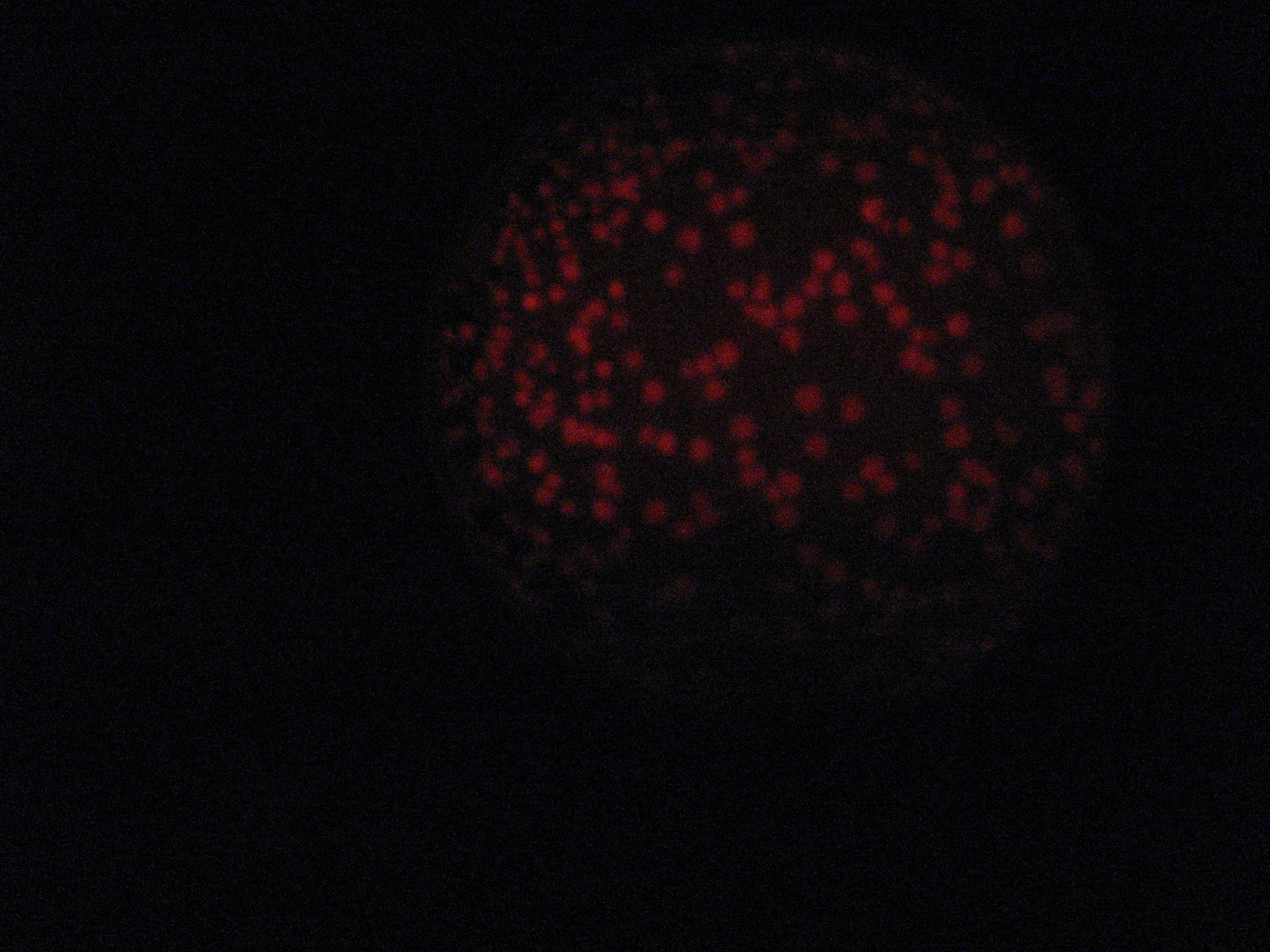 </td> <td>
</td> <td>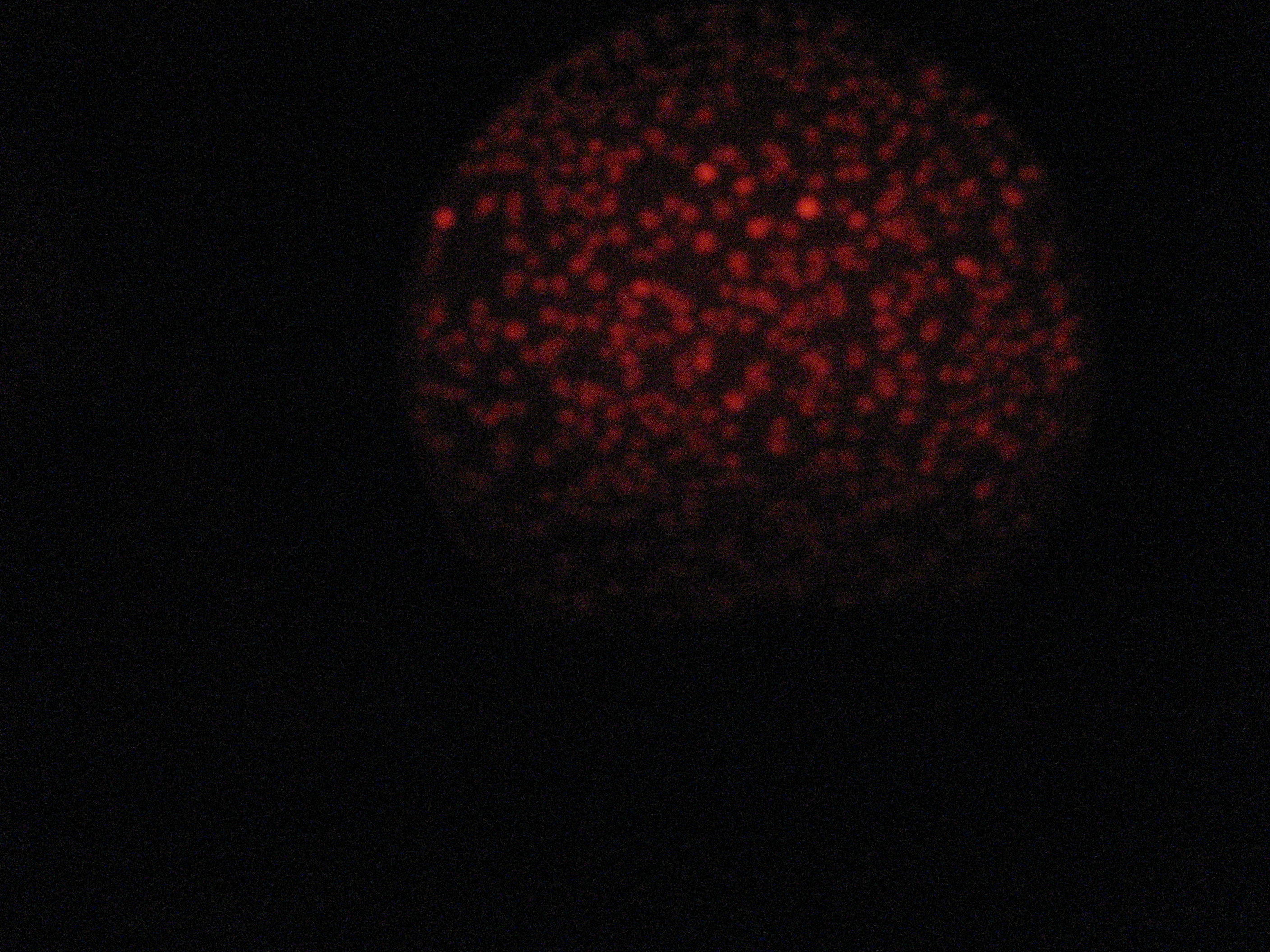 </td>
</tr>
<tr>
<td>1h</td> <td>2h</td> <td>3h</td> <td>4h</td>
<tr>
</td>
</tr>
<tr>
<td>1h</td> <td>2h</td> <td>3h</td> <td>4h</td>
<tr>
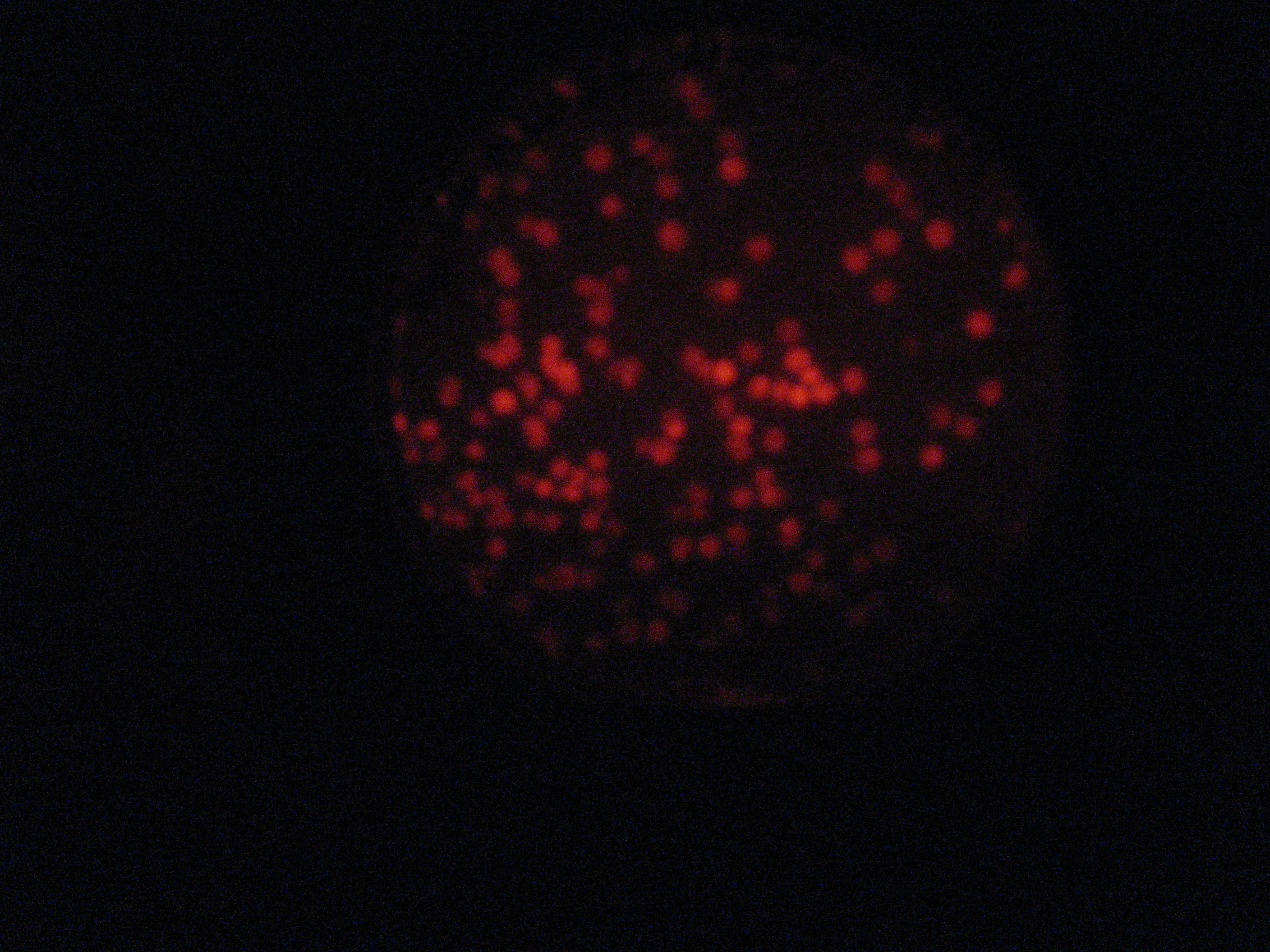
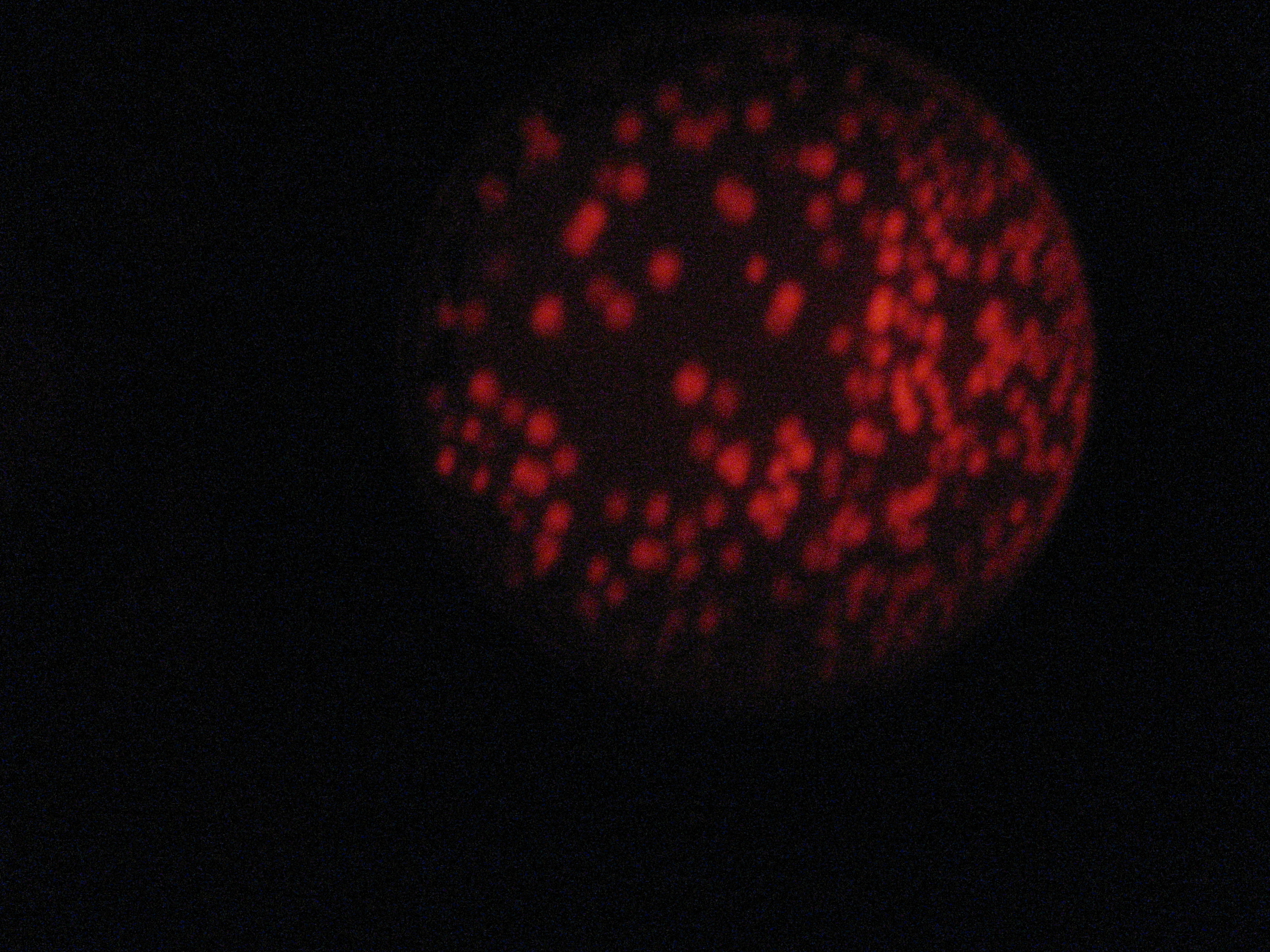
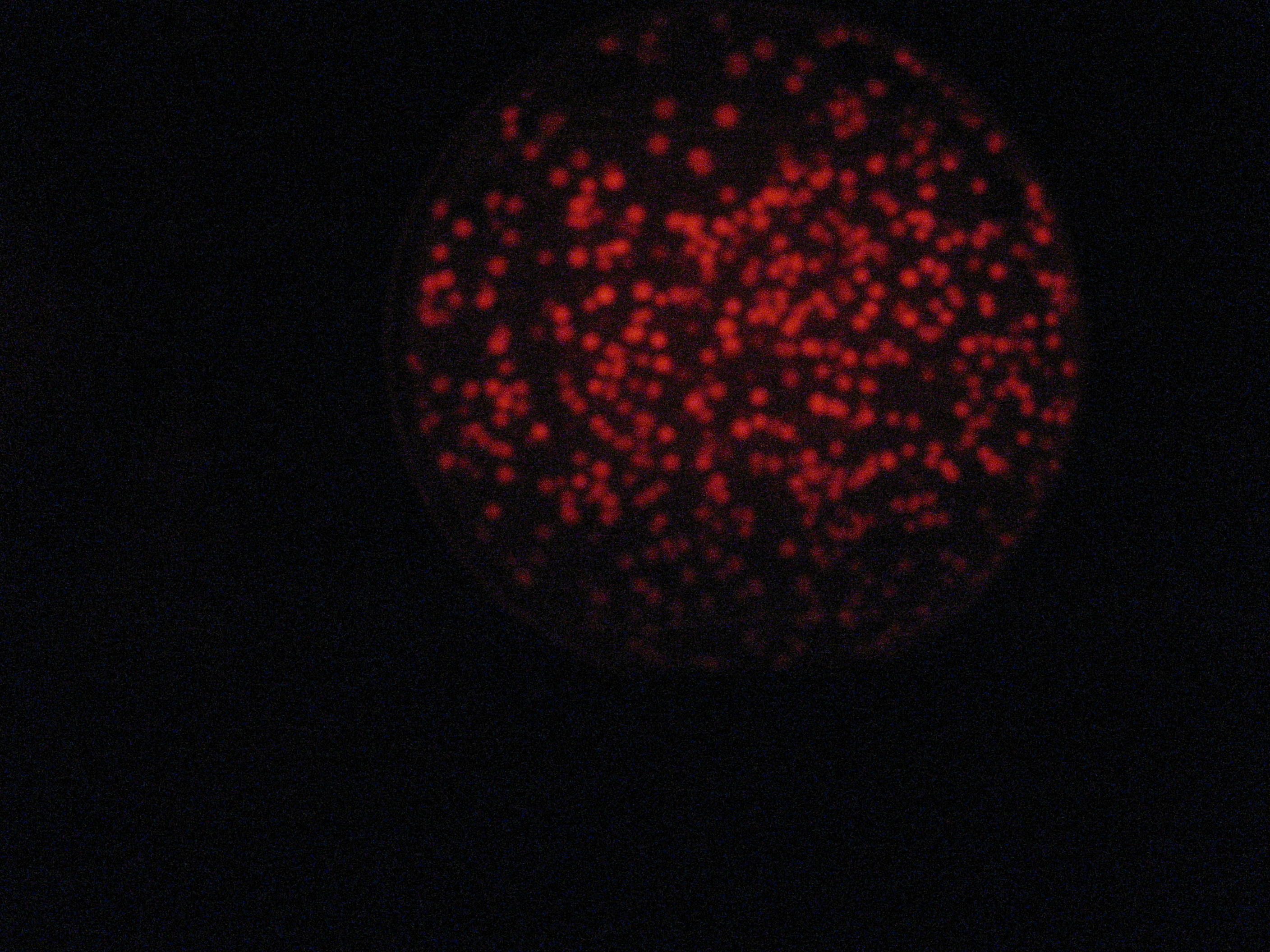 </tr>
<tr>
<td>5h</td> <td>6h</td> <td>7h</td> <td>8h</td>
</tr>
</tr>
<tr>
<td>5h</td> <td>6h</td> <td>7h</td> <td>8h</td>
</tr>
</tr>
- Calculate the recombination rate
- Check the excision of the terminator by sequencing
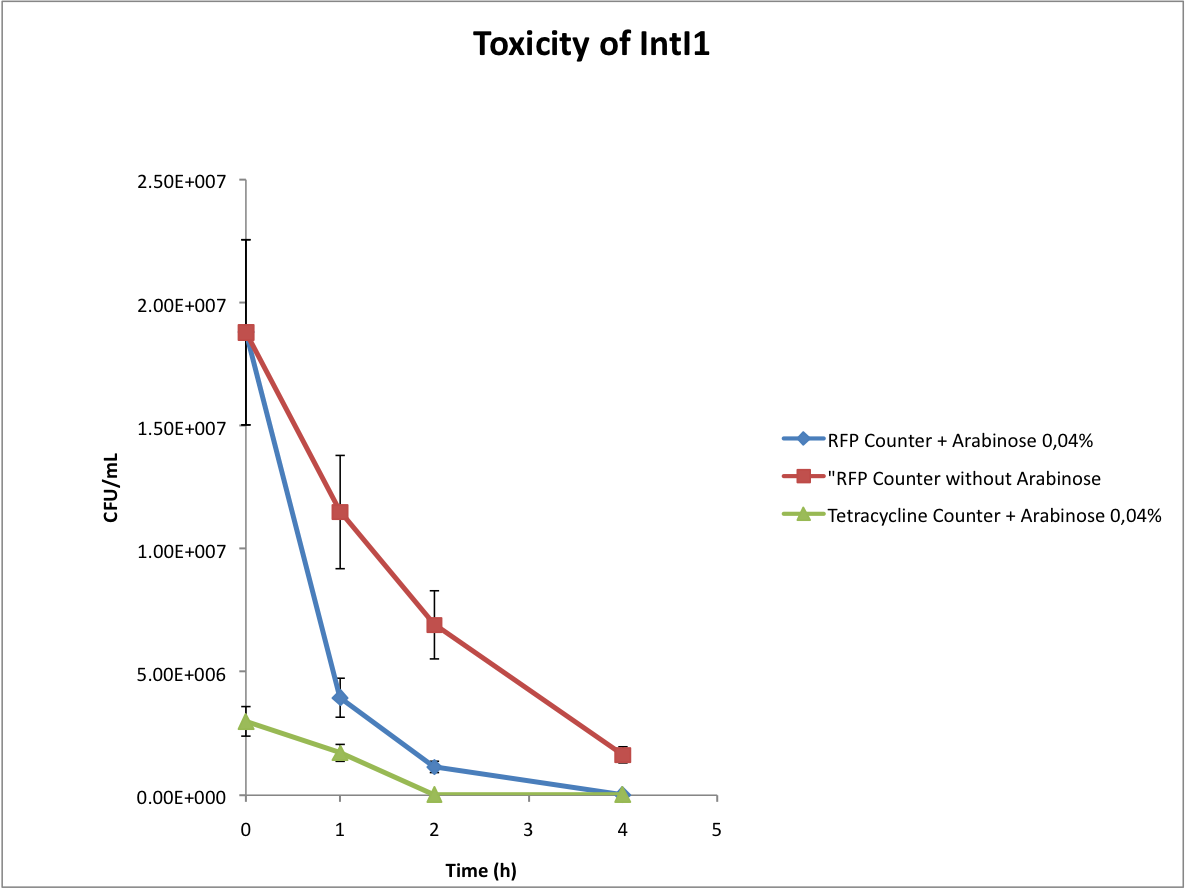
- Verify the toxicity of the integrase
- Test the microfluidic device
 "
"