Team:Caltech/Project
From 2010.igem.org
| Line 48: | Line 48: | ||
The function of the lysis pathway is to release our crosslinking material (i.e. plastic oligomers), exposing it to the extracellular environment. Prior to lysis, the cells will be resuspended in an optically-inactive medium containing our crosslinking reagent, like AIBN, for example. Thus, lysis will release plastic monomers and expose them to AIBN, beginning the crosslinking reaction, possibly with UV initiation. | The function of the lysis pathway is to release our crosslinking material (i.e. plastic oligomers), exposing it to the extracellular environment. Prior to lysis, the cells will be resuspended in an optically-inactive medium containing our crosslinking reagent, like AIBN, for example. Thus, lysis will release plastic monomers and expose them to AIBN, beginning the crosslinking reaction, possibly with UV initiation. | ||
| - | |||
| - | |||
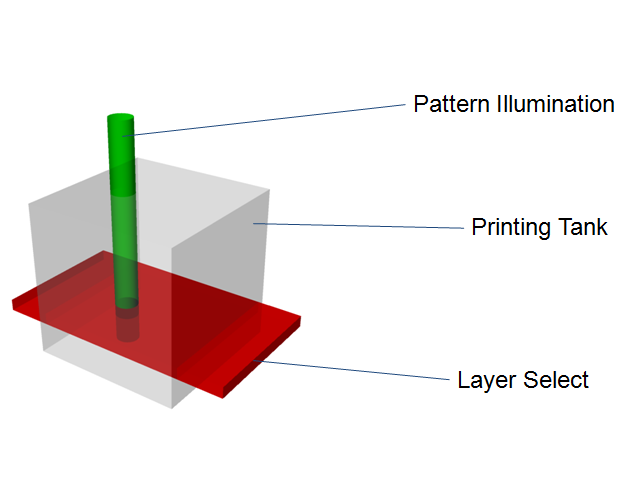
[[Image:LightActivation.png|thumb|right|300px|Simple representation of how pinpoint light activation could be achieved for 3D printing purposes.]] | [[Image:LightActivation.png|thumb|right|300px|Simple representation of how pinpoint light activation could be achieved for 3D printing purposes.]] | ||
| + | |||
| + | Our reaction will be carried out in a 3D reaction volume containing our optically-inactive media and highly-concentrated cells grown overnight to (over-)express our desired product. We will then select a given plane of cells with our infrared plane array, and select the desired shape of cells within that layer using our red light from the third axis (a simple mask will be used to specify the shape). In this way, we can proceed through the entire volume layer-by-layer to create an arbitrary 3D object. Because of our positive feedback loop, we hope to minimize the amount of light activation per layer, which will minimize diffractive activation and the total amount of time required. | ||
After light activation and lysis, we simply wait for the crosslinking reaction to complete, and then rinse the media from the plastic, isolating the desired 3D product. | After light activation and lysis, we simply wait for the crosslinking reaction to complete, and then rinse the media from the plastic, isolating the desired 3D product. | ||
Latest revision as of 00:03, 28 October 2010
|
People
|
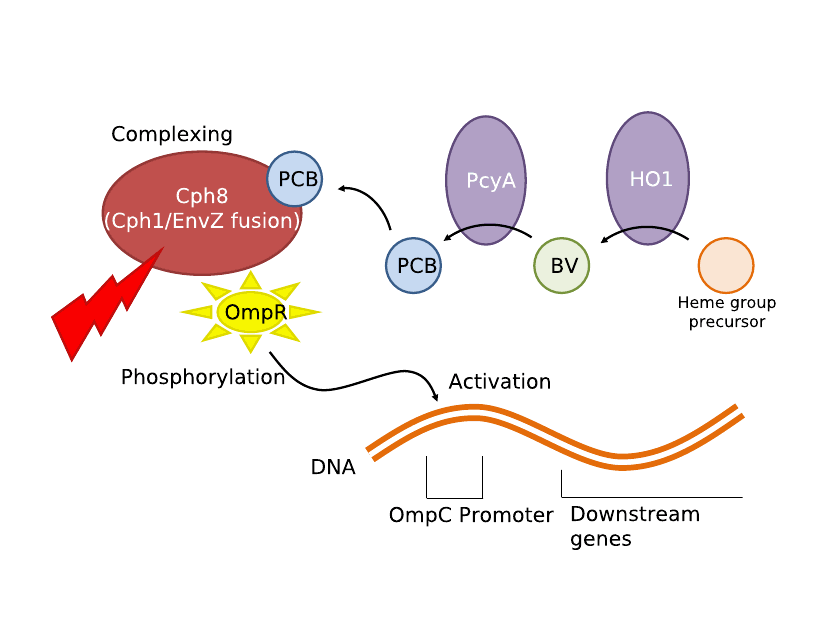
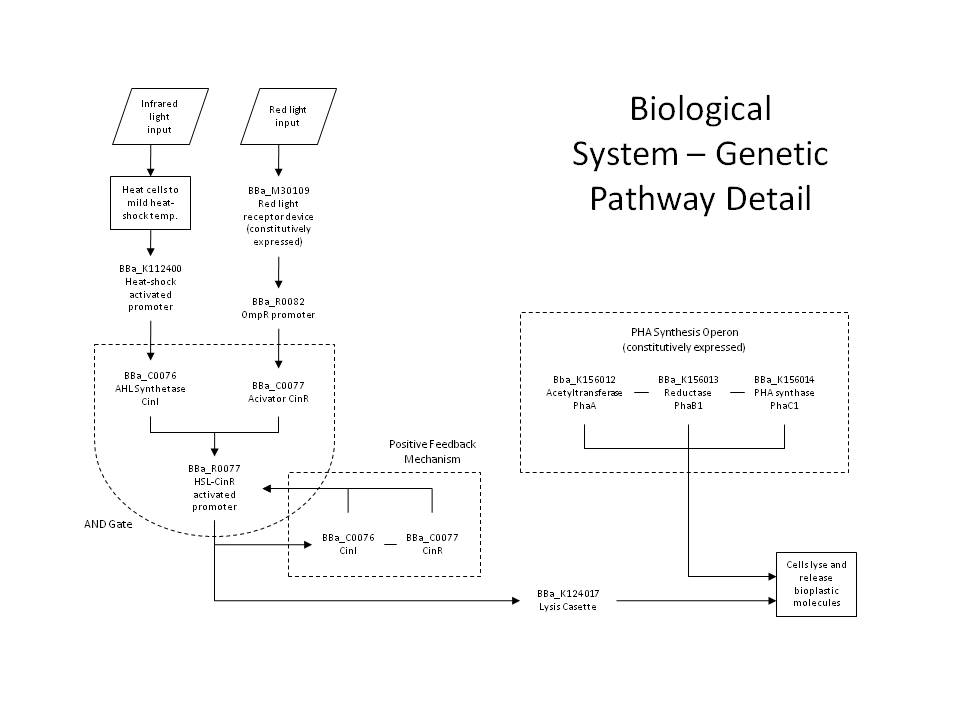
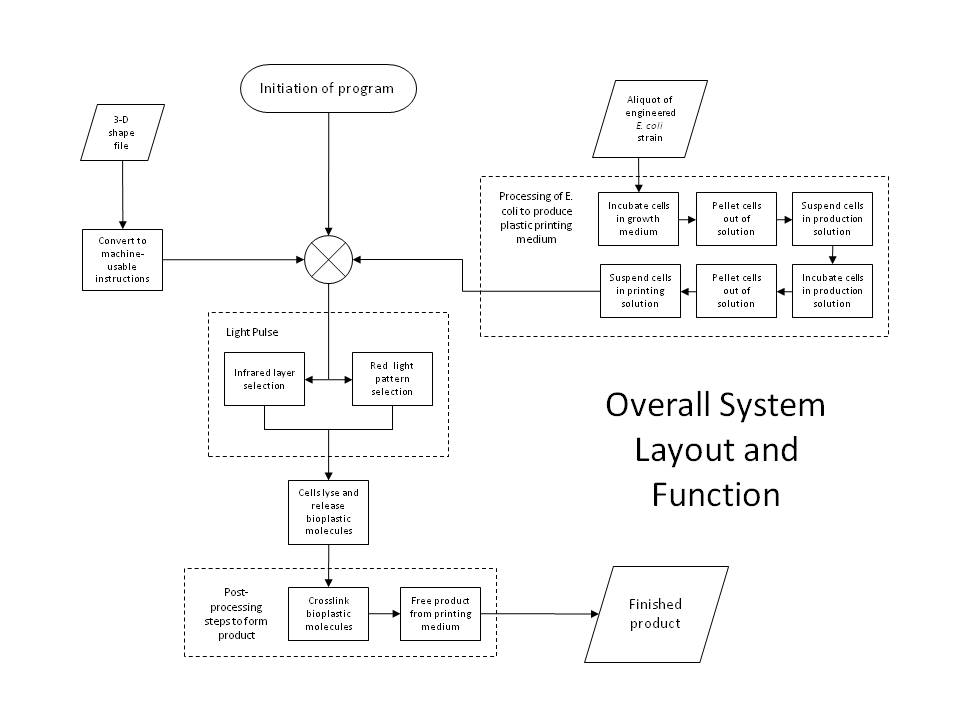
IntroductionWe utilized well-characterized light transduction bricks to induce lysis in our cells. Multiple pathways have been considered: one depends on red (632nm) light to activate an OmpR transcription factor, another depends on infrared light to activate a heat-shock promoter; either of these promoters can be connected to a downstream brick which induces cell lysis. Originally, the red light-induction bricks were used by the UT Austin iGEM team to create bacterial photographs, termed [http://partsregistry.org/Coliroid coliroids]. The heat-shock promoter brick (<partinfo>K112400</partinfo>) was created by UC Berkeley as part of a biological parts suite for bacteria. Similarly, the lysis cassette (<partinfo>K124017</partinfo>) we used was produced by another team – hence the beauty and utility of the Registry: versatility and availability. Ultimately, our team set out to produce a 3D printer with the following specific design constraints: (a) print an arbitrary shape and with reasonable resolution, (b) minimize the cost & time required, and (c) maximize the hardness and utility of the material produced. Light-Induced LysisWe first tried to use the red light transduction system pioneered by Levskaya et al in 2005 [12] and adapted to BioBricks by the UT Austin iGEM team. Upon exposure to 632nm light, the receptor protein would be activated (<partinfo>BBa_I15008</partinfo>, <partinfo>BBa_I15009</partinfo>, <partinfo>BBa_I15010</partinfo>) which then activates a naturally occurring signal protein OmpR, positively regulating a corresponding OmpR promoter (<partinfo>BBa_R0082</partinfo>). Placing this promoter upstream of the lysis cassette (<partinfo>BBa_K124017</partinfo>), completes the simple light-lysis network. Note that the required ancillary receptor genes are contained in a pre-made plasmid (Part <partinfo>BBa_M30109</partinfo>), but should be incorporated into the same plasmid in the final product sequence. Also note that the OmpR system will only work controllably in envZ deficient E. coli; ie in strain CP919 (<partinfo>BBa_V1012</partinfo>).
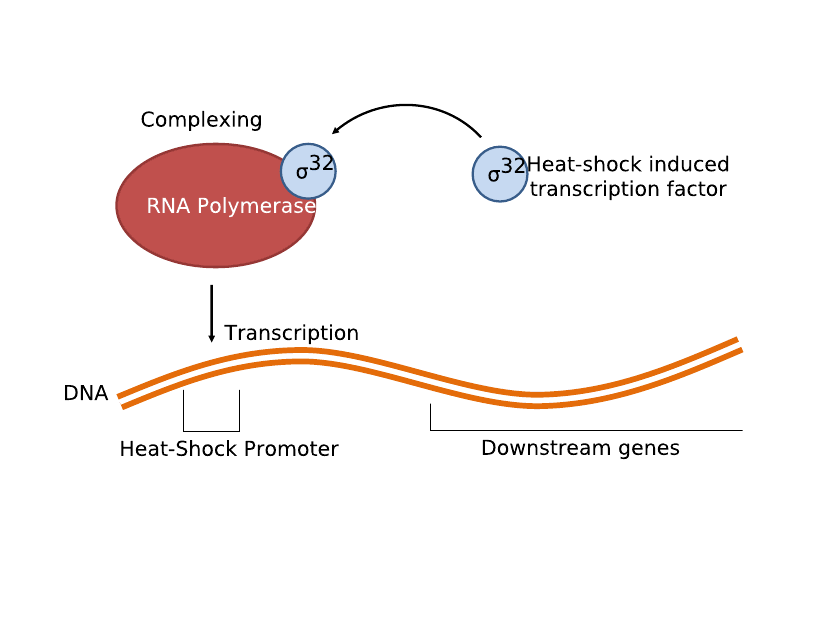
The second and current system involves the heat-shock promoter created by UC Berkeley (<partinfo>BBa_K112400</partinfo>), once again linked to a lysis casette downstream (<partinfo>BBa_K124017</partinfo>) and completing a simple light-lysis pathway. We modified the part to be compatible with the current BBa BioBrick standard and submitted the updated brick as <partinfo>K338001</partinfo>. Infrared radiation would heat up the bacteria a few degrees Celsius, which would trigger the production of σ32 transcription factor, which then complexes with RNA polymerase to activate the heat shock promoter and transcribe the downstream genes.
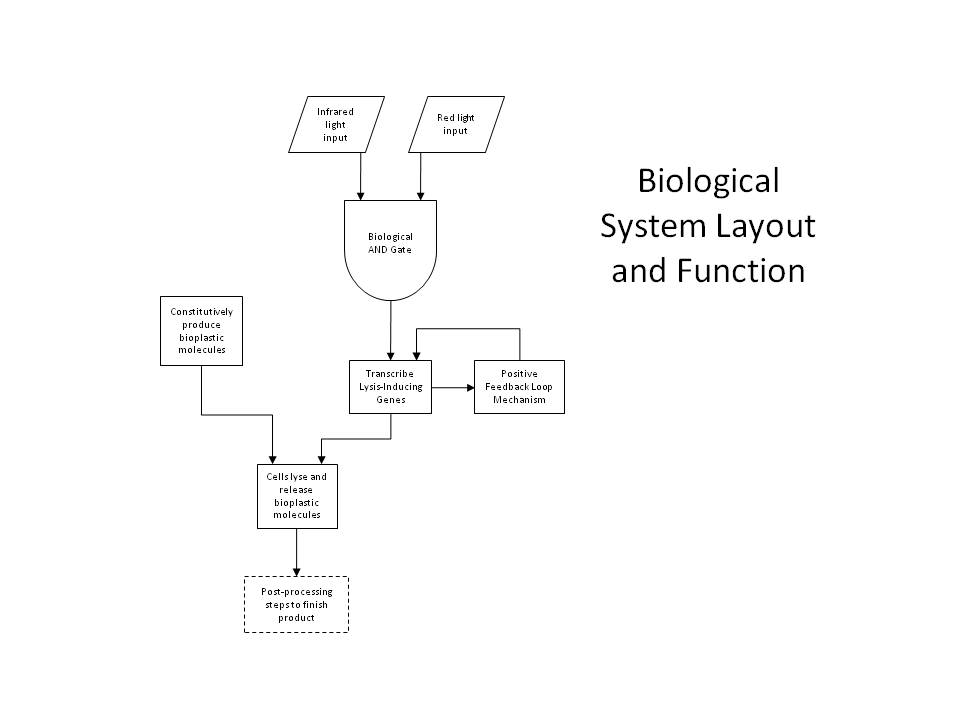
Either simple network is all that is required for two-dimensional (image) selection. We planned to begin by producing flat layers of bioplastic, and work on optimizing the resolution and thickness of each layer, as well as optimizing the properties of the plastic itself by manipulating the crosslinking parameters. However, in order to apply our system to three-dimensional printing, we have designed a more sophisticated gene network, detailed below. Positive Feedback and the AND GateFor increased accuracy and speed, we have designed a more complex gene network to allow dual-wavelength light activation using both red 632nm light and infrared, though the system can be applied generally to any dual-wavelength combination. This way, we can select a given plane within a 3D reaction volume using one wavelength (infrared), and select cells within that plane using the second wavelength (red). The red light activation functions identically with the simpler case, but now the OmpR promoter regulates the production of the CinR activator protein (<partinfo>BBa_C0077</partinfo>). Similarly, the infrared light activating the heat-shock promoter (<partinfo>BBa_K112400</partinfo>), now will cause the production of AHL Synthetase CinI (<partinfo>BBa_C0076</partinfo>). When both CinI and CinR are present in the cell, they form a heterodimer transcription factor which positively regulates the HSL-CinR promoter (<partinfo>BBa_R0077</partinfo>), which then can activate the lysis genes. This HSL-CinR promoter can also be placed upstream of both: (a) the CinI (<partinfo>BBa_C0076</partinfo>) and CinR (<partinfo>BBa_C0077</partinfo>) genes and (b) the lysis cassette (<partinfo>BBa_K124017</partinfo>). In this way, we establish both a positive feedback loop and a strong lysis pathway. The pathway is only activated when the cell has received both red and infrared signals (and has produced both CinI and CinR), comprising the AND gate. This allows us to specify a different lysis profile for each layer in our reaction volume. Because the output of the AND gate activates the further production of the gate's inputs, this also forms an effective positive feedback loop, which ensures that a short pulse of activation signal (light) can be sufficient to produce an abundance of lysis proteins. PrintingOur production cells will not only contain the above gene network, but also genes to constitutively or inducibly create the molecules that we ultimately wish to crosslink. These genes can be placed on a second plasmid, allowing the light activation network to be manipulated independently of that responsible for the printing media. Thus, this system can be considered as a general printing framework, where cell stock can be maintained with only the light-lysis plasmid and the various printing media plasmids can be inserted as necessary, like simple printing cartridges. Any biological material could feasibly be printed with this system, for example including various colors or smells or targeting various properties like hardness or elasticity. Indeed, the system can be considered even more generally still: the dual-wavelength system can be used to easily select any group of cells in a 3D volume, without disturbing the media. Cells can be directed to lyse when activated, as in our case, or can be given any other transcription-based command, perhaps for protein production or regulatory network response. For example, we plan on also adding three PHA (polyhydroxyaldehyde) synthase genes, which will allow our cells to create PHA plastic polymers from soybean oil (specifically: Acetyltransferase phaA (<partinfo>BBa_K156012</partinfo>), Reductase phaB1 (<partinfo>BBa_K156013</partinfo>), PHA synthase phaC1 (<partinfo>BBa_K156014</partinfo>)). To prepare the cells for printing, we will inoculate large liquid cultures and grow them to a high cell density, before transferring them to a medium optimal for the production of plastic polymers, which will activate plastic synthesis. We can then centrifuge the cells to isolate them from their medium and use them for printing. The function of the lysis pathway is to release our crosslinking material (i.e. plastic oligomers), exposing it to the extracellular environment. Prior to lysis, the cells will be resuspended in an optically-inactive medium containing our crosslinking reagent, like AIBN, for example. Thus, lysis will release plastic monomers and expose them to AIBN, beginning the crosslinking reaction, possibly with UV initiation. Our reaction will be carried out in a 3D reaction volume containing our optically-inactive media and highly-concentrated cells grown overnight to (over-)express our desired product. We will then select a given plane of cells with our infrared plane array, and select the desired shape of cells within that layer using our red light from the third axis (a simple mask will be used to specify the shape). In this way, we can proceed through the entire volume layer-by-layer to create an arbitrary 3D object. Because of our positive feedback loop, we hope to minimize the amount of light activation per layer, which will minimize diffractive activation and the total amount of time required. After light activation and lysis, we simply wait for the crosslinking reaction to complete, and then rinse the media from the plastic, isolating the desired 3D product. TransglutaminaseAnother possible crosslinking regime would involve the use of a different crosslinking agent, transglutaminase ([http://en.wikipedia.org/wiki/Transglutaminase TGase]), which is commonly used in the food processing industry to bind meats by crosslinking free amine groups. This enzyme could either be produced in the cell by incorporating it into the lysis pathway (or by secreting it into the extracellular environment) or by direct addition to the lysis medium, like AIBN. This enzyme crosslinks glutamine to lysine residues, and would therefore act on all proteins produced by the cell. Thus, this strategy would work exactly the same as our above system, except AIBN would be replaced by TGase, either produced by the cells or added separately. The plastic production genes would no longer be necessary – rather, we would replace them with genes coding for a high number of glutamine and lysine residues (maximizing the crosslinking potential), and cause these peptides to be heavily produced, inducibly or constitutively. The properties of the resulting hydrogel could be tweaked by adjusting the secondary structure of these over-produced peptides, and their glutamine/lysine distribution and frequency.
|
 "
"