Team:Tokyo Metropolitan/Project/Pattern/Result
From 2010.igem.org
| Line 3: | Line 3: | ||
<style> | <style> | ||
table, tr, td { background-color:transparent;} | table, tr, td { background-color:transparent;} | ||
| - | </style><font size="5"><b>Result</b></font></html> | + | </style> |
| + | <br><font size="5"><b>Result</b></font></html> | ||
| + | |||
=DNA amplifying= | =DNA amplifying= | ||
Latest revision as of 18:33, 27 October 2010

Result
Contents |
DNA amplifying
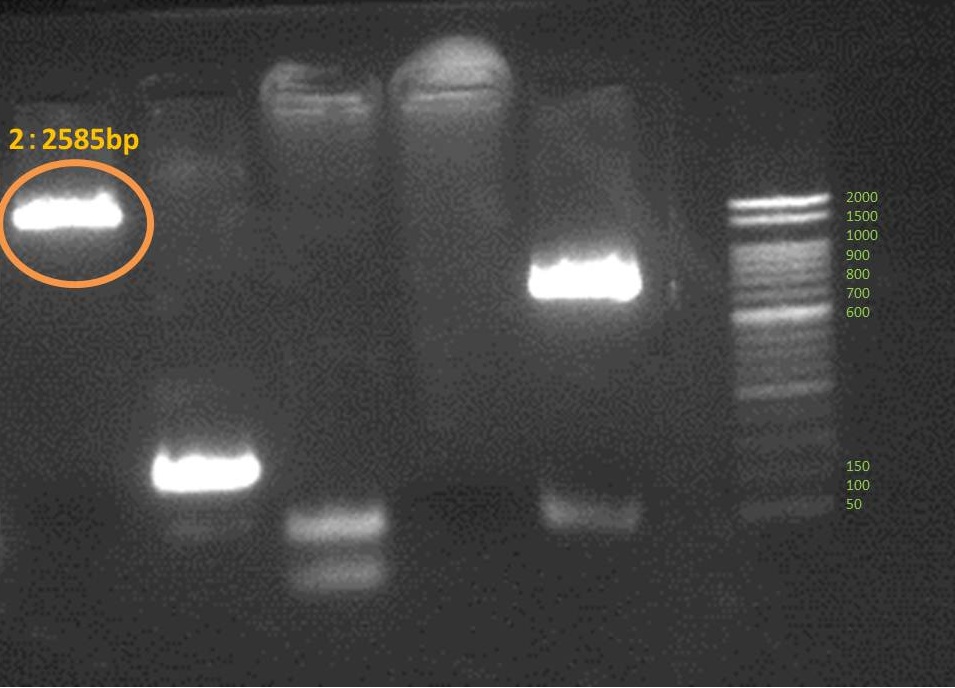
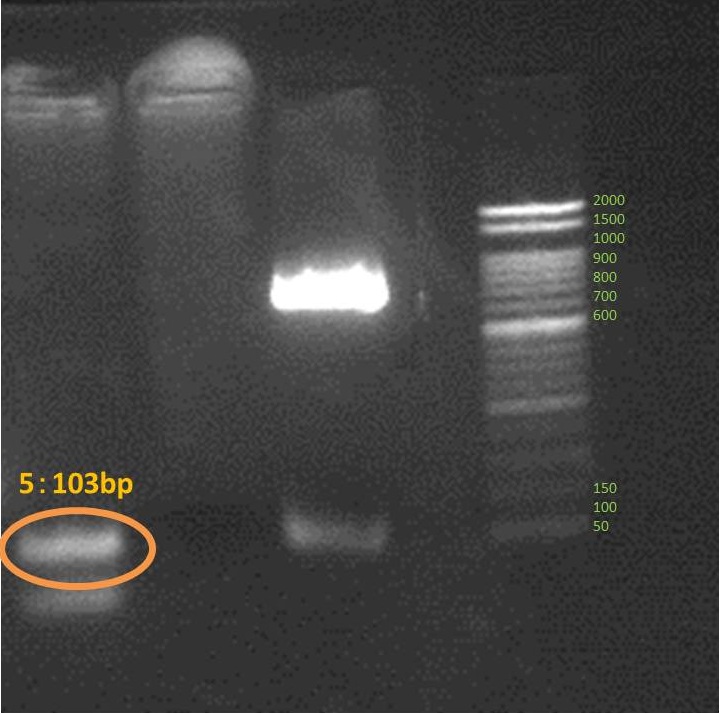
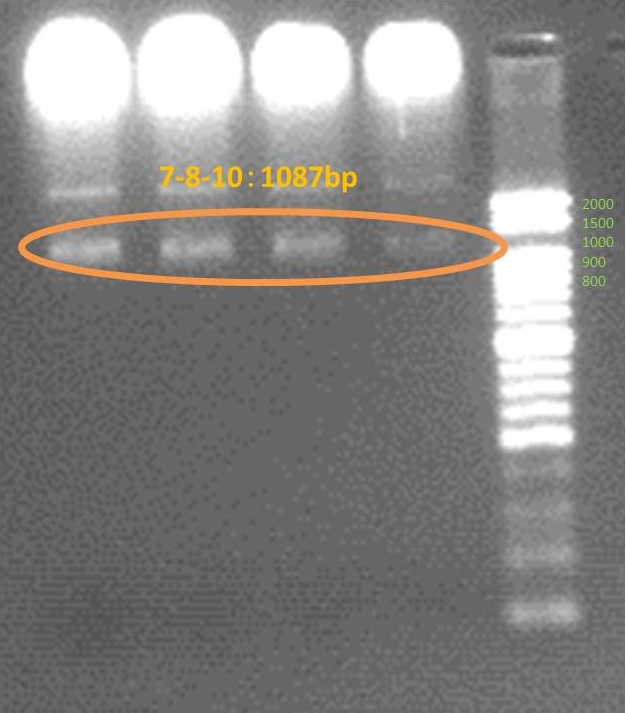
Amplified all genes which are necessary for our projects. These are result of electrophoresis. Each tube number is the same as this gene number.
Activator
We succeed to do PCR 3 DNA fragments which consist Activator. and we could ligate lac promoter to cyaA. but we couldn’t connect mRFP to terminators.
Inhibitor
We succeed to do PCR 5 DNA fragments which consist Inhibitor. but we couldn’t ligate and amplify them.
Device
We succeed to do PCR 3 DNA fragments which consist device to express CRP consecutively. and we could ligate them. but we couldn’t connect them to ruled vector and transform into compitent-cell to cloning. we could detect inserted parts by colony PCR(Insert check) but couldn’t read sequence.
So, unfortunately, we couldn’t reach to reproduct our expected RD model based pattern.
Discussion
So, why we couldn’t complete our project?
- We didn’t have enough skills to do genetic experiments. this is first time that we participate iGEM. and almost all members are ungraduated students. we studied very much about protocols and methods about genetic experiments. but it was very difficult work more than we could imagine.And, because of our low reliance of experimental skill, we can’t examine accuracy of our model. but, its not meaningless experience. we learn and got skills through this project. In next year, we ensure that we can do more good experiments and results.
____
 "
"