Team:Monash Australia/Modelling
From 2010.igem.org
(→Model 2) |
(→Kinetic modelling of the ethylene generator) |
||
| Line 11: | Line 11: | ||
| - | The program tinkercell was used to model the synthesis of ethylene in our E.coli cells. Where, in addition to the qualitative representation (figure 2 & 4) the program allowed, simulation of the model could be exercised inorder to measure the flux of metabolites – with respect to time - in the system given the input of values pertaining to substrate/enzyme concentration, and reaction rates were inputted. In addition to | + | The program tinkercell was used to model the synthesis of ethylene in our E.coli cells. Where, in addition to the qualitative representation (figure 2 & 4) the program allowed, simulation of the model could be exercised inorder to measure the flux of metabolites – with respect to time - in the system given the input of values pertaining to substrate/enzyme concentration, and reaction rates were inputted. Figure 2 or Model 1 was designed to include fixed concentrations at a steady state ratio of 1:1:1. This design ensured that ballpark ethylene figures were obtained and all parameters and enzyme rates were in correct proportions to achieve a reasonable output. |
| + | |||
| + | |||
| + | In addition to model 1, quantitative data in silico corresponding to mRNA, protein translation, transcription and degradation were implimented into a more complex design. As a result, more realistic outputs of ethylene could be found. This design, model 2 could then be used to comapre and contrast actual ethylene values obtained via experimental design. Hence the effiency of our E.coli cells could then be calculated. Depending on the effiency value, certainn factors and variables could be changed in order to attempt to reach our tinkercell output. | ||
| + | |||
| + | Another use of the tinker cell model is that it will allow us to understand the pathway in which we choose to take our project. Low concnetrations of ethylene will allow ethylene to be used in a plant comunicating systems. However if ethylene present in high concentrations then the ethylene could be used in plastic productions. Therefore the tinkercell system gives us an indication of which direction the Monash iGEM team can take in future years | ||
==<H2> Aims of the Kinetic Modelling </H2>== | ==<H2> Aims of the Kinetic Modelling </H2>== | ||
Revision as of 11:43, 27 October 2010

|
|
|||
Contents |
Introduction
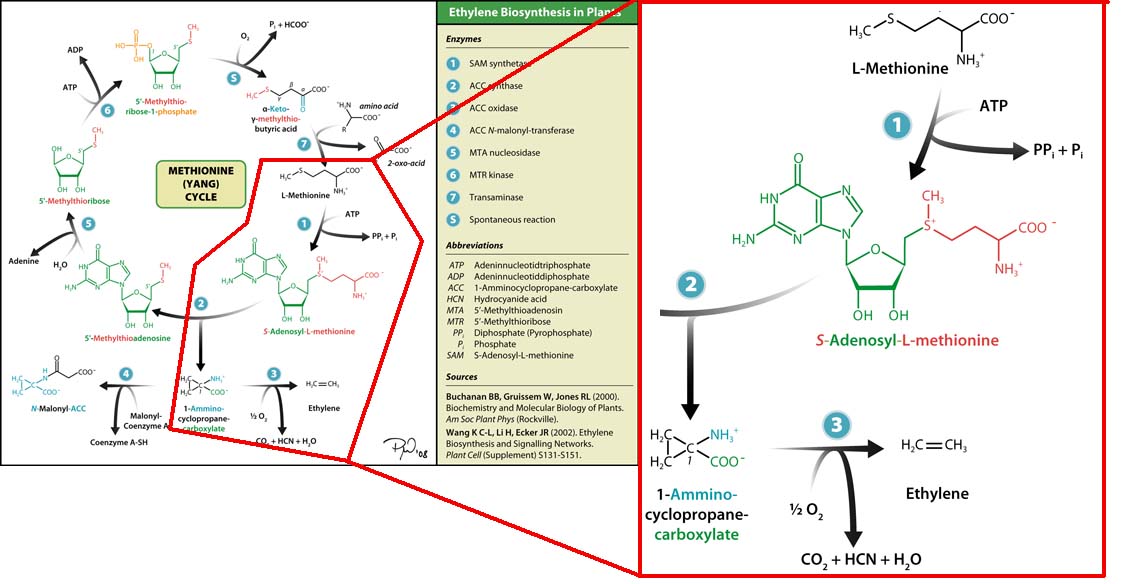
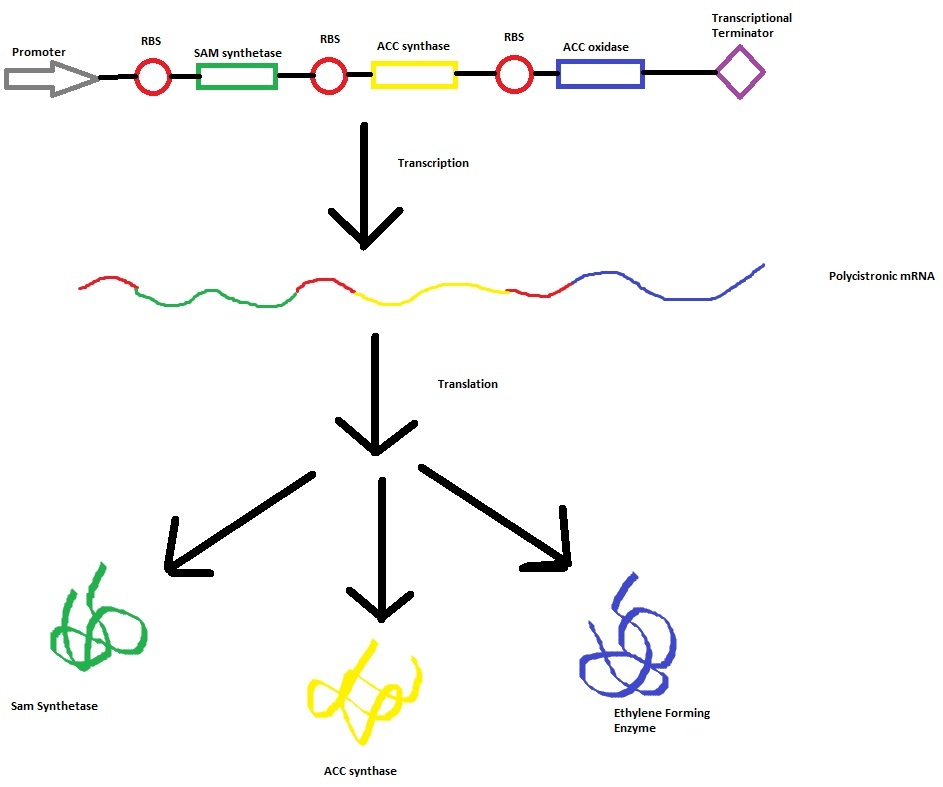
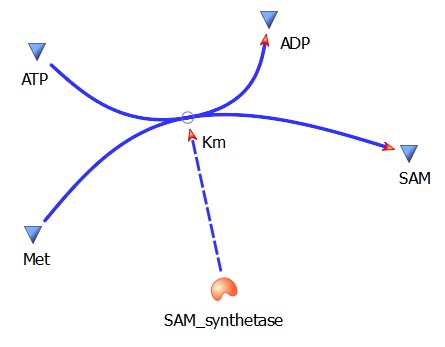
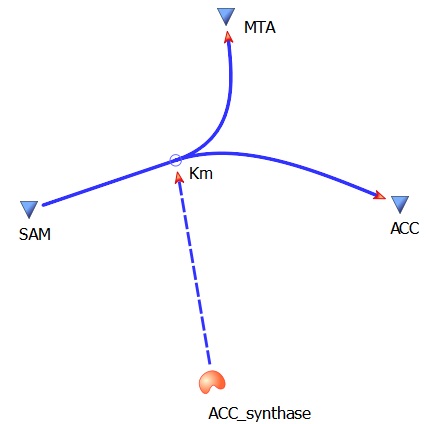
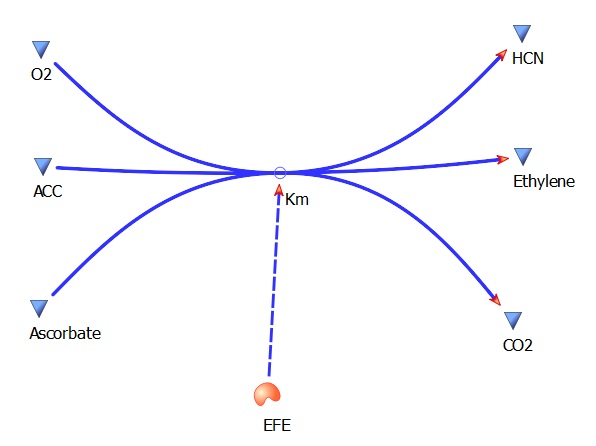
The three key enzymes we require are outlined in the image Figure 1 (left), SAM synthase, ACC synthase and ACC oxidase. SAM synthase converts methionine into S-Adenosyl-L-Methionine (SAM), using ATP for an adensoyl group. The second step involves ACC synthase, which cleaves the amino butyrate from SAM, releasing 1-aminocyclopropane-1-carboxylic acid (ACC). Released ACC is then processed by ACC Oxidase which converts ACC to ethylene by cleaving the carboxylic acid off as carbon dioxide and its neighboring carbon with the amino group as cyanide gas. By using such a system to produce ethyene gas we can potentially reduce costs involved with current production methods by reducing temperature requirements by 30 fold.
Kinetic modelling of the ethylene generator
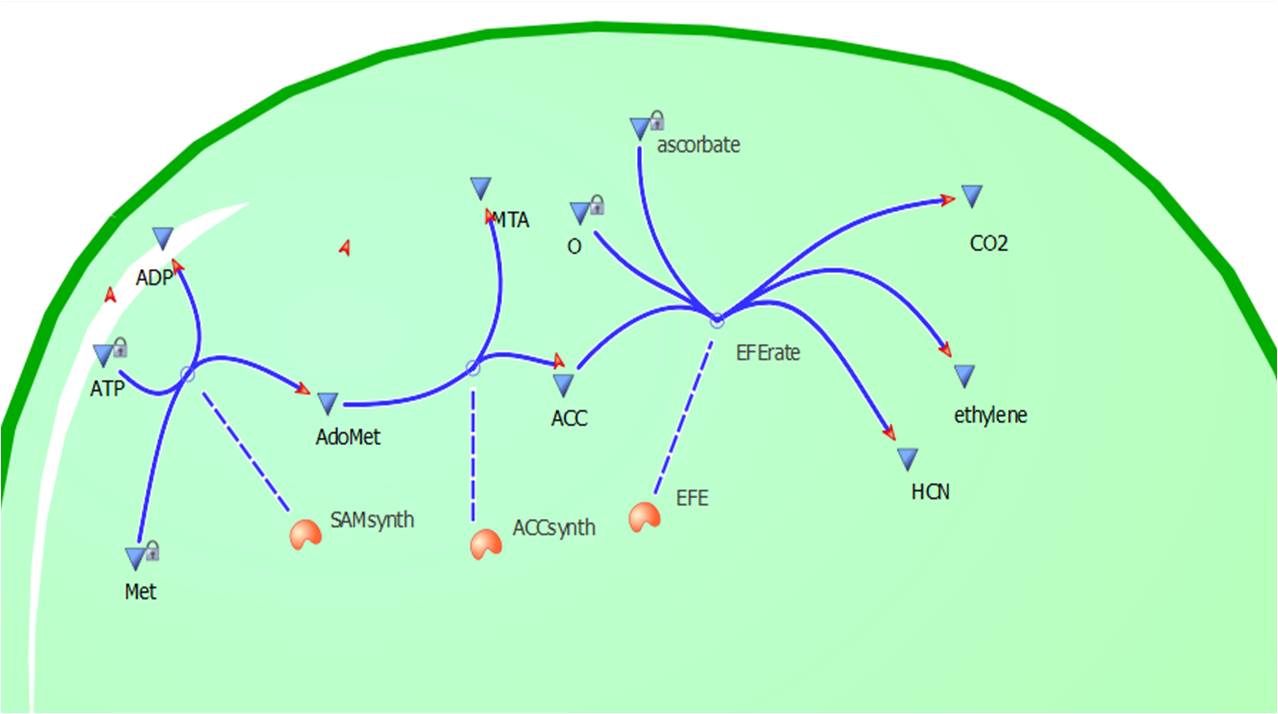
The program tinkercell was used to model the synthesis of ethylene in our E.coli cells. Where, in addition to the qualitative representation (figure 2 & 4) the program allowed, simulation of the model could be exercised inorder to measure the flux of metabolites – with respect to time - in the system given the input of values pertaining to substrate/enzyme concentration, and reaction rates were inputted. Figure 2 or Model 1 was designed to include fixed concentrations at a steady state ratio of 1:1:1. This design ensured that ballpark ethylene figures were obtained and all parameters and enzyme rates were in correct proportions to achieve a reasonable output.
In addition to model 1, quantitative data in silico corresponding to mRNA, protein translation, transcription and degradation were implimented into a more complex design. As a result, more realistic outputs of ethylene could be found. This design, model 2 could then be used to comapre and contrast actual ethylene values obtained via experimental design. Hence the effiency of our E.coli cells could then be calculated. Depending on the effiency value, certainn factors and variables could be changed in order to attempt to reach our tinkercell output.
Another use of the tinker cell model is that it will allow us to understand the pathway in which we choose to take our project. Low concnetrations of ethylene will allow ethylene to be used in a plant comunicating systems. However if ethylene present in high concentrations then the ethylene could be used in plastic productions. Therefore the tinkercell system gives us an indication of which direction the Monash iGEM team can take in future years
Aims of the Kinetic Modelling
-To determine the maximum output of ethylene -To estimate HCN levels and assess potential damage to cell
-Start with a simple model and progress to a more complex one. This allows us to more accurately predict ethylene production
Model 1
Introduction to Model 1
This relatively simple model - as represented in figure 2 - was designed so to be the starting point of our kinetic modelling. This uncomplicated system could then be used to obtain an ethylene output. This design comprised only of three enzymes and lacked the transcription and translation stages. The enzymes invloved were SAM synthetase, ACC synthase and ACC oxidase - also known as EFE (ethylene forming enzyme). These enzymes were present at fixed concentrations (10, 100, 1000 and 3000 uM) and in a 1:1:1 ratio. This ensured that the model was plain yet ultimately obtained a resonable yet simple ethylene output. Therefore with fixed enzyme concentrations the rate at which ethylene is produced depends on the Vmax of the specific enzymes.
This specific rate is defined by the Michaelis-Menton Equation:
kcat*[substrate]*[enzyme]
Rate = -------------------------
km + [substrate]
Assumptions for Model 1
- A 1:1:1 steady state of enzymes.- No product inhibition.
-Substrates (ATP, L-Methionine etc) kept at a constant value due to homeostatic mechanisms in E.coli.
- The best value (km, kcat etc) was always chosen to ensure maximum yield of ethylene.
Parameters and Values used
Met concentration - 150 uMATP concentration - 9600 uM
O2 concentration - 442 uM
SAMsynth Km(Met)- 92 uM
ACCsynth Km(AdoMet/SAM)- 37 uM
EFE Km(ACC)- 12 uM
SAMsynth turnover rate (kcat)- 1.5 per second
ACCsynth turnover rate (kcat)- 18 per second
EFE turnover rate (kcat) - 5.9 per second
Graphs and results
Using Tinkercell a graphical representation of 'Model 1' was generated (Figure 3) using deterministic stimulation. In this figure a graphical representation of the rates - generated internally by Tinkercell's mathematical alogarithms - of ethylene production are shown for each fixed enzyme concentration assumed. As a result not only is production of ethylene shown to be increasing linearly with time, but at a greater rate with increasing concentration of enzyme (SAM synthetase, ACC synthase, EFE) used.
-
Model 2
Model 2 explores the more complicated version of our kinetic modelling. It aims to incorporate the transcription and translation rates of the cell into the final design. This calculation begins to become more difficult due to problomatic degradation, transcription and translation rates. These values were obtained from the [http://www.ebi.ac.uk/biomodels-main/BIOMD0000000012 Elowitz repressilator model]. Of the many sources browsed it was determined that this source seemed to be the most accurate and hence its values were used. Therefore the transcription and translation rates of mRNA and proteins constantly changes the final concentration of the three enzymes. This changing concentration of enzymes then produces ethylene at a more reliable rate. As a result this model is expected to produce a more accurate representation of the ethylene producing pathway.
Elowitz Degradation Values
mRNA degradation rate - 0.00577 mRNA per second Enzyme degradation rate - 0.001155 enzymes per second R0011 PoPs or transcription rate - 0.5 mRNA per second Translation rate -0.117 enzymes per secondGraphs and results
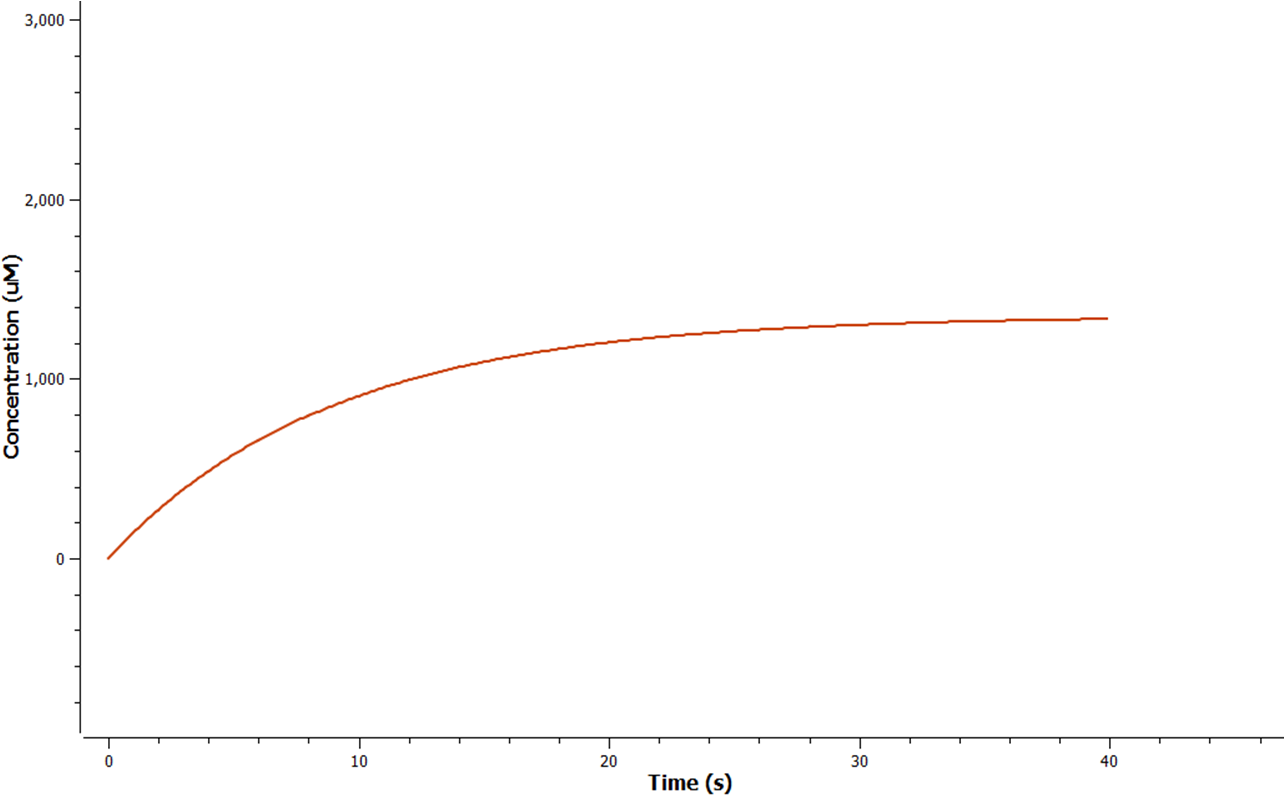
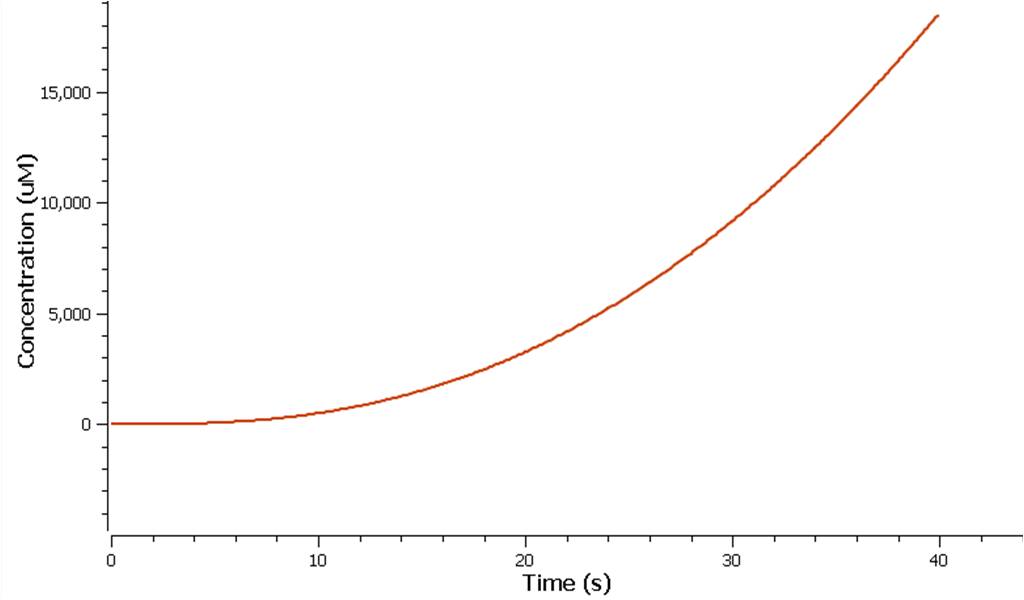
The rate at which mRNA is produced can be seen to start off at a rapid rate. However as time goes on the mRNA can be seen to reach a steady state.
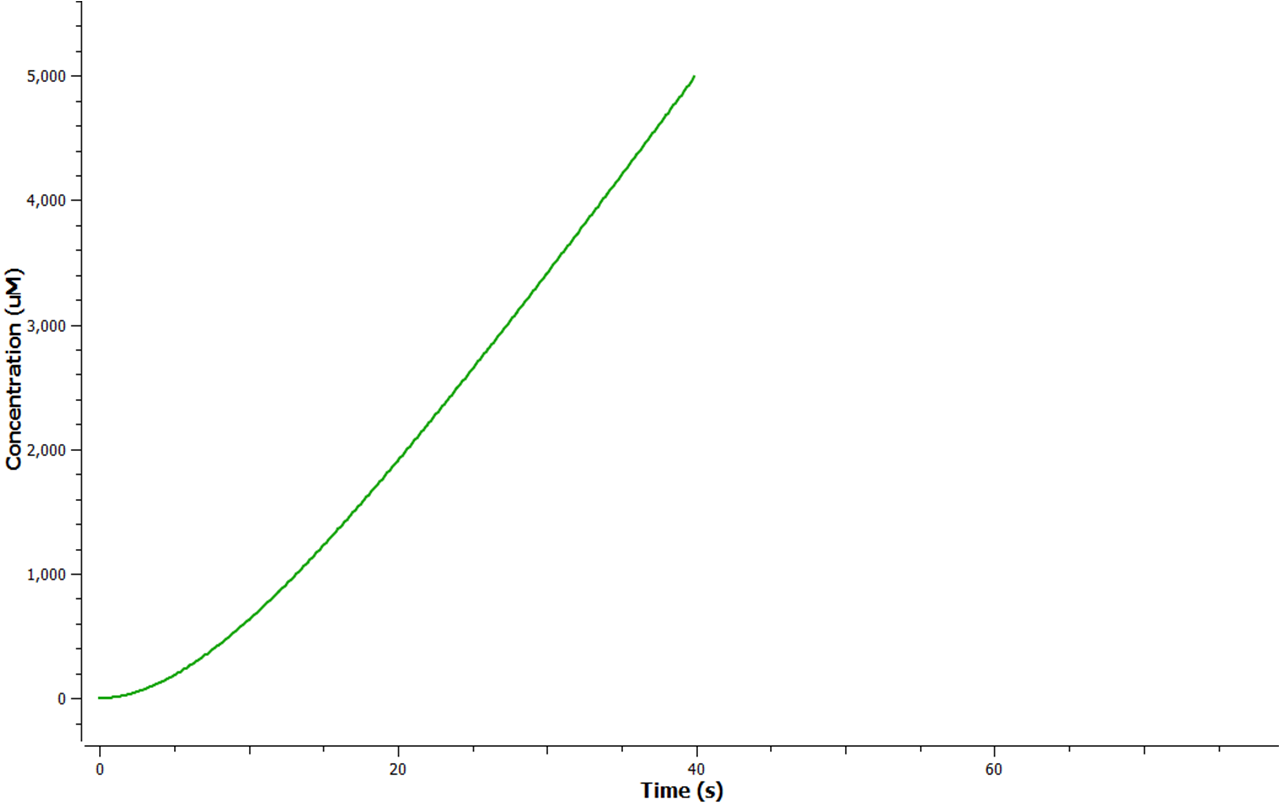
The production of ethylene in this graph looks realitively accurate, however due to the confunding aspects of excess enzyme production, it is unclear if this graph truly represents the total ethylene output. However it can be assumed that the first few seconds accurately portray the ethylene output.
Construct --> mRNA --> enzymes (SAMsynth, ACCS, EFE)
Met --> SAM (AdoMet)
SAM --> ACC
ACC --> Ethylene
More to come soon!
 "
"