Team:Peking/Project/Bioabsorbent/MBPExpression
From 2010.igem.org

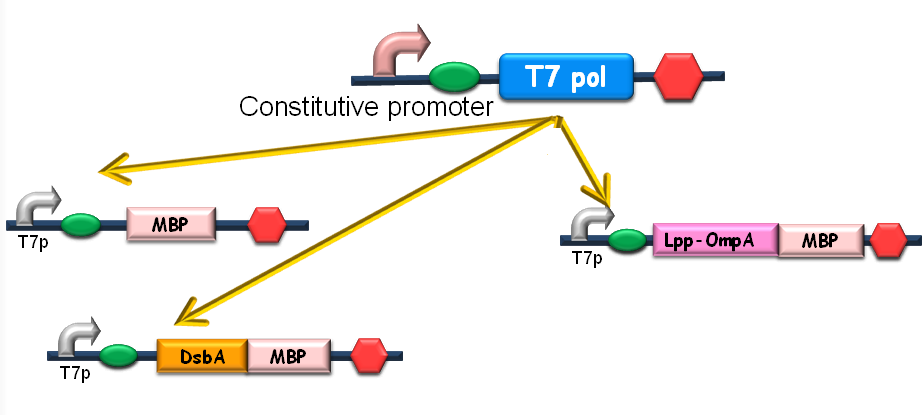
Since our ultimate goal is to design a high-performance and less energy-consuming bioabsorbent, the MBP will be an excellent candidate for the absorbent effector. MBP was then fused with DsbA, a periplasmic translocated protein with a signal sequence at its N-terminal, and OmpA, a membrane protein, to construct periplasmic MBP and surface displayed MBP, in order to maximize the bacterial capability of mercury binding. Finally, mercury MBP was constitutively expressed on surface, periplasm and cytosol of E.coli cells via carefully designed genetic circuits, to guarantee the maximum of Hg absorption (Fig 1).
Figure1. The genetic circuit we designed to guarantee the maximum of Hg absorption. The production of T7 RNA polymerase is constitutive. T7 polymerases will active high rating transcription at T7 promoters. Thus Hg (II) will be highly effectively accumulated by substantial amount of MBPs which are translocated to cytosol, periplasm and cell surface of the bacteria.
Cytoplasmic Expression of MBP
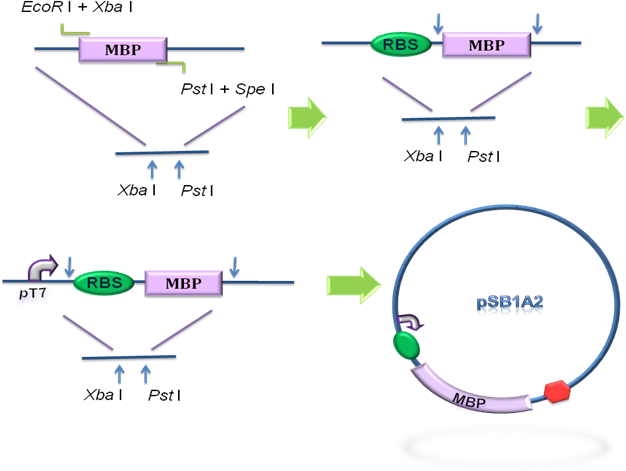
To design the module of cytosol expression, MBP in pSB3K3 was then to coloned into pSB1A2, prefixed by T7 promoter and RBS BBa_B0034 (Fig 2), verified by DNA sequencing.
Fig 2. Construction of the cytosol expression module. MBP was cloned into the pSB1A2 step by step, prefixed by T7 promoter and RBS.
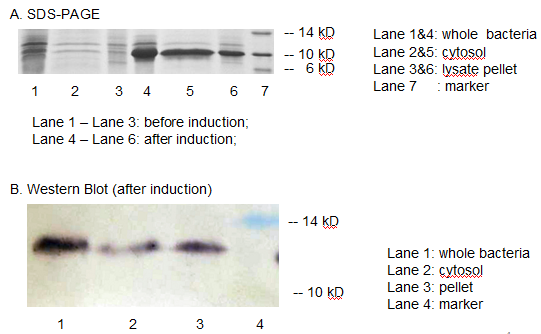
The resulting construct, T7 promoter + BBa_B0034+ MBP was transformed into BL21 (DE3) competent cells. The expression of MBP has been verified by SDS-PAGE and western blotting. (Fig 3)
Fig 3. The expression of MBP has been verified by SDS-PAGE and Western Blotting. IPTG was used as the inducer. (A) Overexpression band at about 10 KD was detected. (B) Positive band of the expected molecular weight could be detected in the cytosol
We detected overexpression band at about 10 kD, which was likely to be MBP. Then we fused a his-tag to our target protein and conducted western blotting to further verify their expression. Positive band of the expected molecular weight could be detected in the cytosol, which confirmed expression and localization of the target protein. We can also indicate from the SDS-PAGE that there are indeed a large number of MBPs existing in cytosol, ready to bind mercury ions.
METAL BINDING PEPTIDE (MBP) CONSTRUCTION
To achieve the goal of making a high performance MBP, we constructed a single polypeptide consisting of two dimerization helixes and metal binding loops of MerR, to form an antiparallel coiled coil MBP mimicking the dimerized metal binding domains of the wild-type as described in Fig 2. We amplified the N-terminal and C-terminal of MBP directly from full length MerR by PCR, and then cloned them into the backbone together in one step (Fig. 4).
A
B
Figure 3. MBP Construction procedure. A: Standard part. B. Expression detection part.
To be specific, the entire coding region of the MBP for standard part was amplified by PCR from full length MerR with two pairs of primers. Two of these primers encoded a three-residue bridge, SSG, which does not occur in MerR and was added to afford some flexibility in the loop connecting the two dimerization helix (fig. 1). The two PCR products were digested with EcoR I / BamH I, or BamH I / Pst I and cloned into EcoR I / Pst I -digested pSB1K3 in one step (fig. 3A), which was verified by DNA sequencing.
Based on the same strategy, MBP-His6 was constructed by using two different pairs of primers, which is used for MBP expression test by western blot. The two PCR products were digested with Nde I / BamH I, or BamH I / Xho I and cloned into Nde I / Xho I -digested pET 21a, which contains a region encoding six histines, in one step to construct pET 21a – MBP (fig. 3B), which was verified by DNA sequencing.
Reference
1.Chen, S., and D. B. Wilson. 1997. Construction and characterization of Escherichia coli genetically engineered for bioremediation of Hg2-contaminated environments. Appl. Environ. Microbiol. 63:2442–2445.
2.Chen, S., and D. B. Wilson. 1997. Genetic engineering of bacteria and their potential for Hg2 bioremediation. Biodegradation 8:97–103.
3.Chen, S., and D. B. Wilson. 1997. Construction and characterization of Escherichia coli genetically engineered for bioremediation of Hg2-contaminated environments. Appl. Environ. Microbiol. 63:2442–2445.
4.Luirink, J., and B. Dobberstein. 1994. Mammalian and Escherichia coli signal recognition particles. Mol. Microbiol. 11:9–13.
5.Debarbieux, L., and J. Beckwith. 1998. The reductive enzyme thioredoxin acts as an oxidant when it is exported to the Escherichia coli periplasm. Proc.Natl. Acad. Sci. USA. 95:10751–10756.
6.Francisco, J. A., Earhart, C. F. & Georgiou, G. (1992). Transport and anchoring of beta-lactamase to the external surface of Escherichia coli. Proc Natl Acad Sci U S A 89, 2713–2717.
7.Francisco, J. A., Campbell, R., Iverson, B. L. & Georgiou, G. (1993). Production and fluorescence-activated cell sorting of Escherichia coli expressing a function antibody fragment on the external surface. Proc Natl Acad Sci U S A 90, 10444–10448
8.Yamaguchi, K., Yu, F. & Inouye, M. (1988) Cell 53, 423-432.
9.Jie Qin,Lingyun Song,Hassan Brim, Michael J. Daly and Anne O. Summers(2006) Hg(II) sequestration and protection by the MerR metal-binding domain(MBD).Microbiology 15, 709–719
 "
"