Team:USTC Software/html5
From 2010.igem.org



- Overview
- Features
-
Fun and Function
MoDeL: Modeling Database Language
Bring Biological Modeling to the Next Level
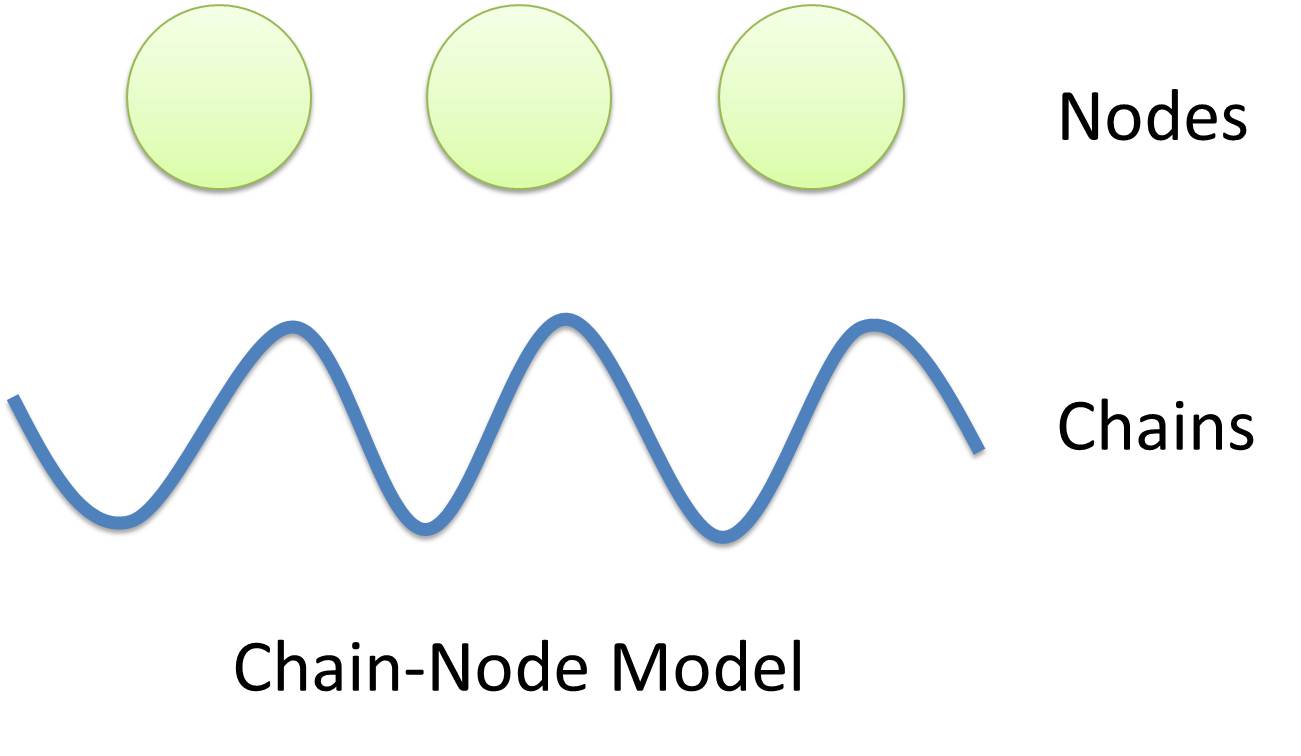
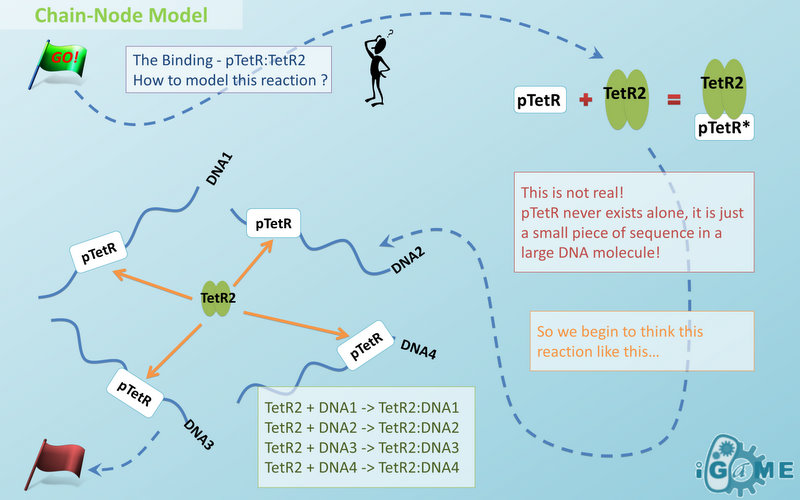
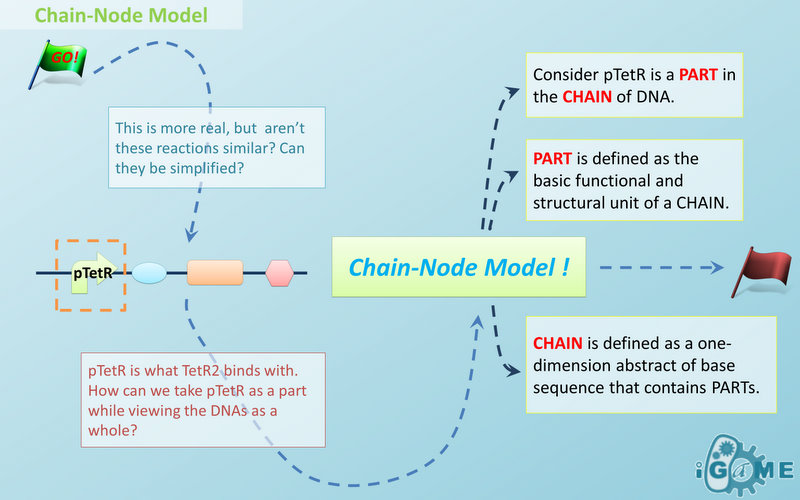
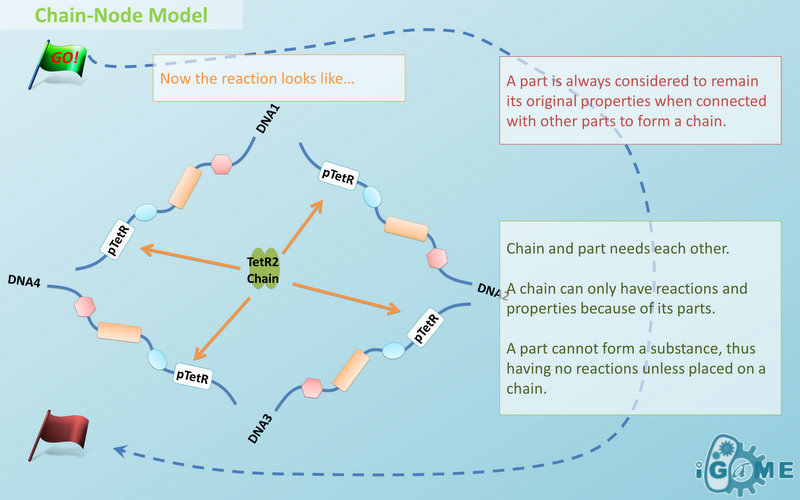
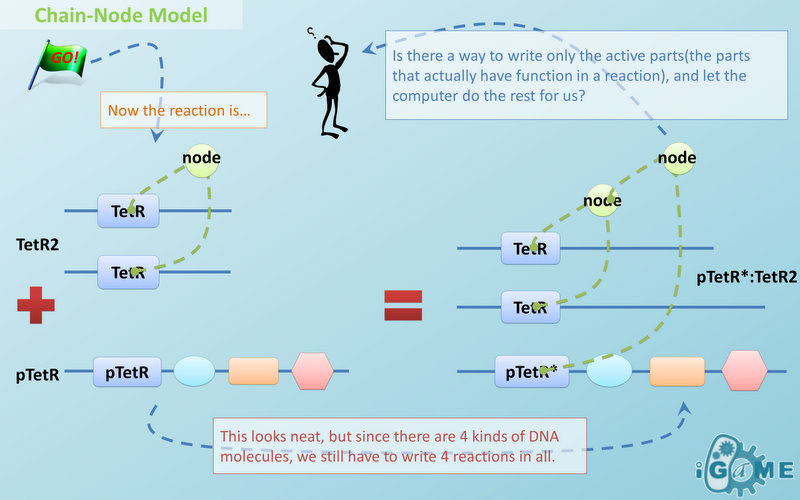
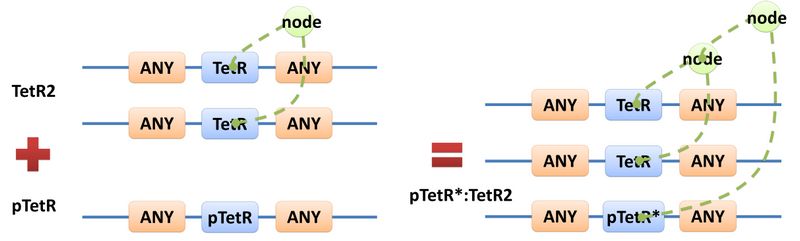
Chain-Node Model (Figure. 1) is a brand new Complex Modeling Concept incorporating its detailed structure description with universal applicability. Instead of treating complex as a whole while ignoring their basic composition and structure, Chain-Node Model view complex as a construction of it basic Parts. Just as its name implies, our model includes two components: Chain and Node. As a correspondence to natural polymer chains, each Chain consists of an arrangement of its basic unit, Part, whose concept has been greatly extended and includes but not limited to Biobrick Parts. The Node component is not a natural correspondence but an abstract concept to describe binding states of two or more parts: each binding will create a Node. The abstract nodes may continue to bind with other parts or nodes to form a tree structure. However, parts or nodes in bound states are not allowed to bind again. With help of chains and nodes, it is possible to model any complex with arbitrary architecture. Simple and inaccurate modeling of biological process could not keep pace with the development of synthetic biology and undoubtedly, our Chain-Node model provides a possible solution to the imbalance. A simple example, tetR dimer, is shown to illustrate our simple modeling idea (做一个tetR2的模型放在右边做为配图,否则右边太空了). It has two chains with each containing only one part, tetR. Dimerization of tetR will create a node to indicate the bound state of two parts. To explain more clearly, bound parts are also considered as nodes so that in this example, all nodes are organized in a tree structure, which includes two children (leaf) nodes and one parent node. We will conform to this convention in our wiki.
To know more, users are suggested to read this One-Minute Introduction to have an intuitive idea and deeper understanding of our modeling system.
Modeling with Templates
Modeling manually of biological system is widely used for synthetic biology modeling but it requires an overall understanding of the biological network. It is difficult for even professionals to provide such large amount of data. The underlying reason making modeling so difficult is that the data provided manually are redundant because different reactions may occur through different mechanisms. Based on this. we are always seeking feasible ways to implement our automatic modeling idea. The automatic does not mean modeling without any information provided, but however, there indeed exists a minimal data set to enable the automation.
Similar to C++ programming language, we introduce templates to allow generic description of species and reactions of a certain pattern. It is essential to implement our automatic modeling idea since
to share the same
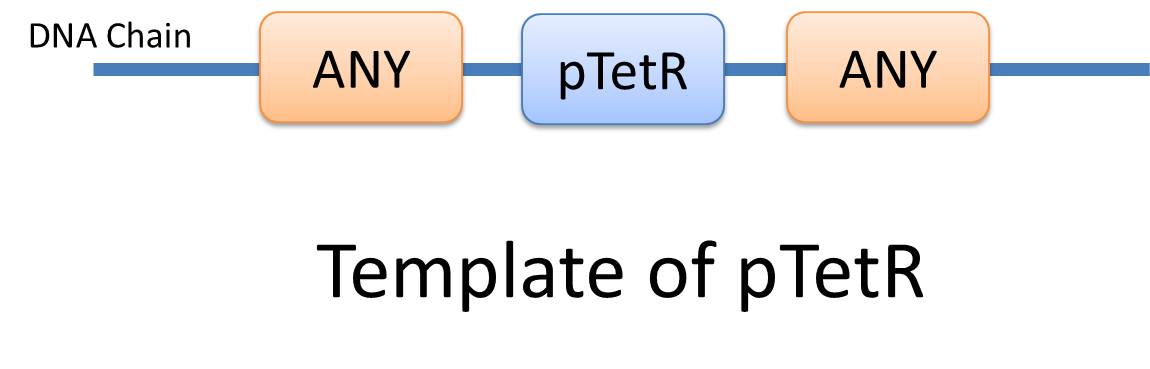
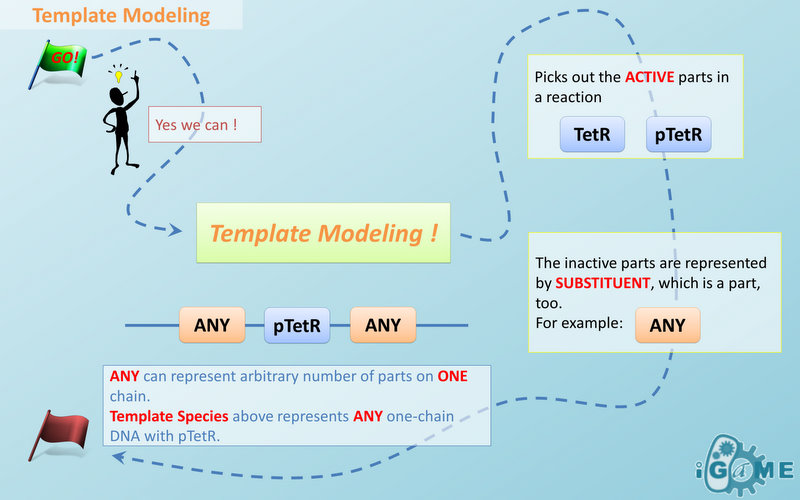
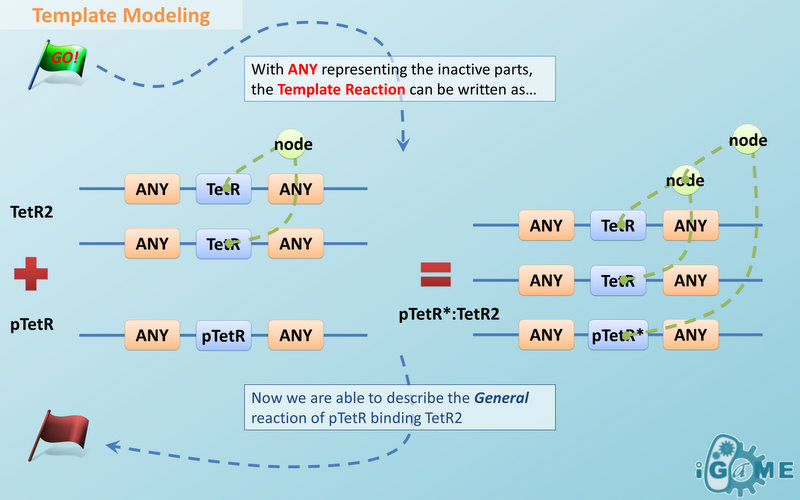
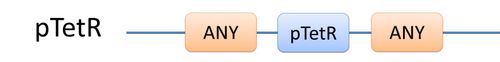
Automatic modeling of biological system, Similar reactions happen for similar reasons. For example, the repression of pTetR promoter by TetR dimer can happen on many different DNA molecules with pTetR, regardless of what other biobricks is constructed on the DNA. With this thought, we add the part of Substituent, which can replace inactive parts of Species in Reactions, making it more general, and actually turning it into a reaction template. Having substituent, a pTetR TetR binding reaction is re-interpreted as the binding reaction of ANY DNA with pTetR(Figure 2) with ANY protein with TetR-dimer binding site. Modeling with templates allows us to describe reactions of new complex even without rewrite the reactions and species in database.
Automatic Modeling Database Language
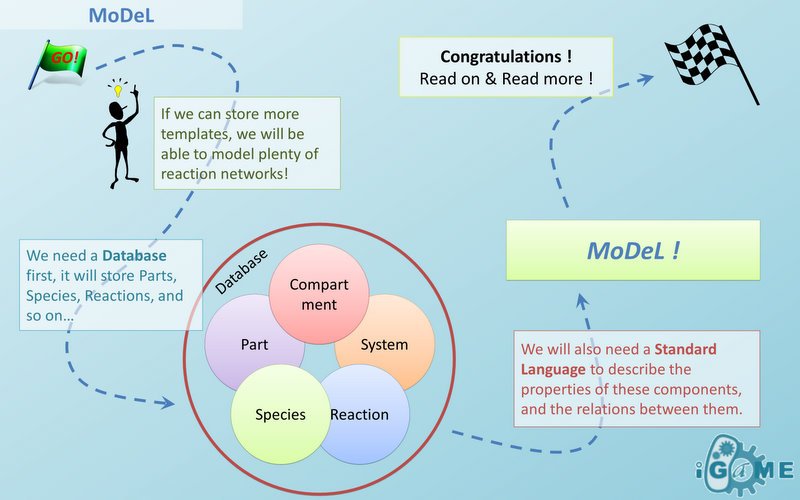
We use a database to store all the information we need in modeling. In order to realize automatic modeling, we construct the database in unified format and make it machine-readable. Every component of database has its specified attributes and values, which makes the format of the database a unique yet standard database language. We call it MoDeL: Modeling Database Language by picking out characters from three words. MoDeL is based on XML language, which makes it flexible and extensible. For more specifications of MoDeL, click here.
- User Interface
-
User-Friendly Design
Input
Output
- MoDeL
-
One-Minute Introduction
You are about to begin an exciting journey of the main ideas in MoDeL. For each picture, start from the green flag, follow the arrows, and finally you will reach the destination! Read on from the first step or click one of the three steps!
Chain-Node Model -> Template Modeling -> Automatic Modeling Database Language
Chain-Node Model
Template Modeling
Automatic Modeling Database Language
- Modeling
-
Introduction of MoDeL based Automatic Modeling Algorithm
Development of Modeling Database Language (MoDeL) aims to provide a platform that enables separation of work between modeling and data constructing. Users could obtain dynamic behaviors of various quantities and other properties of a biological system as the output of iGame while they know nothing about details of modeling and the reaction network. Previous modeling and simulation tools require users to construct the network themselves, which is very hard even for professional researchers. iGame has no such requirements and users are only required to input data about the initial environmental condition of system. Hence, modeling is a task that is performed to find all reactions and species in the system.
To complete the final network from initial environment conditions, we first break up the task to several parts:
(1) Setup initial conditions and environmental parameters read from the input file.
(2) Species produced are inserted into a list in the order of time. For each species, we search the database to find template species which could be matched to this species in structural pattern. If not found, then go for the next species.
(3) For each template species we found, we continue to search template reactions containing this template species as a reactant or modifier or both. Only forward or reverse reaction is handled, never for both.
(4) For each template reaction found, we search in the species list for possible species which could be instances of templates of other reactants and modifiers. If not found, then go for next reaction.
(5) For each possible combination of reactants and modifiers, if the reaction parameter conditions are satisfied, we generate the structure of products based on the transfer table and structural templates of products. And then add this new reaction to the reaction list.
(6) Go to step 2 until the end of the species list.
In the following, we will focus on the step (2), (3) and (5). Our algorithms are totally different with that of other software tools since we design them for the purpose of proving the feasibility of MoDeL for synthetic biology. They are kind of complicated because the complexity of biological systems require so.
What are special reactions and How to handle with them?
Transcription and translation reactions are not supported by MoDeL reaction definitions; they are handled specially in the core program. We use pseudo first-order reaction to model both transcription and translation reactions (RNA polymerase are ignored). The rate constant k is the value of forwardPromoterEfficiency/reversePromoterEfficiency for transcriptions and forwardRbsEfficiency/reverseRbsEfficiency for translations defined for each part in the database. A transcription will not end at a terminator with termination efficiency less than 100% (values are stored in attribute forwardTerminatorEfficiency/reverseTerminatorEfficiency): RNA polymerase will pass over and continue transcription. For translation reactions, the prerequisite condition is that the part next to RBS along the 5' to 3' direction must have non-zero attribute value forwardStartCodon/reverseStartCodon. Opposite to transcriptions, translation reactions would stop if one part with non-zero attribute value forwardStopCodon/reverseStopCodon is encountered.
What are differences from single compartment to multi-compartment?
Consider an example first. AHL molecules outside diffuse into E.coli cells with rate k1, while AHL molecules inside E.coli cells diffuse outward and enter into the bio-reactor with rate k2. We assume diffusion reactions are all driven by concentration gradient.
Chain Sorting and Weighting Scheme
liaochen@mail.ustc.edu.cn
Chains in our Chain-Node Model should not have any order since there are no natural ways to sort them. One chain is not preferred to another, since in a complex of multiple chains, which are connected via binding interaction of binding bites on their own, all chains are equivalent. However, a chain-sorting program is a prerequisite for implementations of many other functions, such as comparison of two Chain-Node structures, or pattern matching of template species to those with definite structure.
Sorting is always performed under some rules. Since there are no natural options, we design one to generate the unique order for a list of chains. The rule we developed is something like that for comparing two strings: Compare first the partReference and partType defined in database of the first part of two chains in alphabet order and if equal, then compare the next until there are differences or the end of either chain. Meanwhile, a part in bound and unbound state(这里多了半句?)If unicode_c of each chain is different with others, then we could sort the chain list by comparing their unicode_c (just the same with comparison of two strings). However, it is not always so: chains with same unicode_c may be equivalent or not. It is due to the mark asterisk *. Unicode_c knows where are the binding sites, but it does not know the binding details of that part. In other words, unicode_c could not solely determines the order of the chains since it lacks of information of the trees defined in our Chain-Node model. The forest (set of trees) in Chain-Node Model describes the binding details of parts in chains. We need to find something like unicode_c to represent the essence of the associated trees. Our weighting scheme will be first introduced below.
Assume the chains are sorted already. Each part will be assigned a weight based on its position in the chain list. The weight of the jth part on the ith chain is (i,j). For example, the first part on the first chain posses a weight (0,0) (in C style, counting number from zero). After assignment of parts' weight, we turn to weights of the nodes in trees. Since leaf nodes in trees are actually the representation of parts in bond state on chains, the weight of each leaf node could be inherited from the weight of its corresponding part. For parent node, its weight equals to the maximum of weights of all its children nodes (we define a weight (i, j) is greater than another (i',j') if i < i' or i = i' while j < j'). The recursive definition of tree nodes' weight make it possible to sort the forest. For convenience, we add a virtual node ROOT, whose weight value could also be obtained under the general rule, as the root of all trees in the forest and make a conversion from forest to tree. The tree structure could be uniquely determined by recursively sorting all children of each nodes by their weights.
Back to the question above, if two chains have the same unicode_c, which should be arranged ahead? The answer lies in the exchange of them. The exchange will change the parts' weight on both chains and thus the weight of some nodes of the trees. The tree structure (with virtual root) is also possible to change. If the tree structure is not changed, the two chains are equivalent, which means that it makes no difference to arrange either chain in front of the other. If the tree structure changes, we must find some ways to determine which tree structure is preferred and based on this to arrange the two chains with equal unicode_c. We know each tree could be uniquely converted to a binary tree by taking the first child of each node as the left child and its siblings as its right child. The Huffman algorithm could be used to generate the Huffman code of each node in the binary tree. Since the Huffman code is actually a representation of the path from current node to the root, we can restore the tree structure from the set of Huffman codes of all leaf nodes. Hence, the Huffman codes set of leaf nodes could uniquely represent a tree structure. We could generate a string, unicode_t for tree structure by splicing the Huffman codes and their weight of all leaf nodes sorted by their weights and adding a asterisk between two adjacent codes, for example:[000:0:1][001:1:3][010:2:3][011:2:4]
Based on this idea, we could obtain the tree unicode_t for any possible order by exchanging equivalent chains and choose the order which make the minimum unicode_t value. By checking the unicode_t value of permutation, chains are grouped in a number of equivalent classes, which contains one or more chains. Chains within each equivalent classes are equivalent: exchanging order of any two chains will not change the tree structure.
It is easy to compare two Chain-Model structures with the help of chain sorting. We only need to compare unicode_c value of chains in the chain list and unicode_t value generated under that order. If two structures are equal, their chain list and tree structure must be equal since the chains order is unique in our chain sorting method. If two structures have same unicode_c value of each chain and unicode_t value, they must be same structures since we could uniquely generate a Chain-Node structure from the strings.How to find species with specific structural pattern of a template one?
We assume that if a combination of species in the species list could be structurally matched to reactants and modifiers templates defined within a reaction, they are possible to react with each other in the way as the reaction template describes. Hence, it is important to find such matchings between species in the species list and template ones defined in the database. We will first give the basic idea and then some modifications.
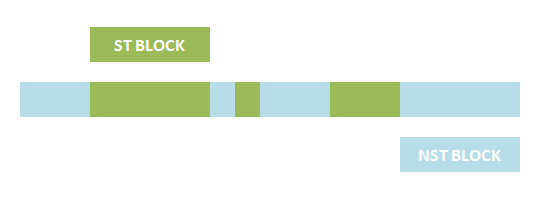
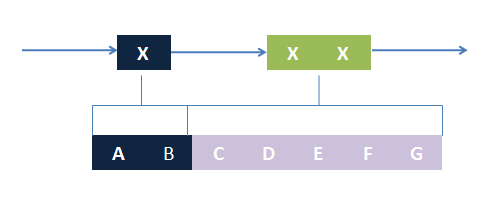
The basic idea is simple. We first consider the structural pattern matching between one chain and a chain template. There are two kind of parts on a chain template: substituent-type part (ST) and non-substituent-type part (NST). The whole template chain could be separated into blocks of ST parts and NST parts. Then we could break down the task to the matching of each small block. For each NST type block, we find all its matching regions on the chain to be matched, there may be more than one region. Each of the combinations of all NST blocks forms a NST frame and restricts the matching regions of ST blocks. Therefore, the minimal task is to find matchings of each ST part block within a restricted region on the chain to be matched. At the present version of MoDeL, there are 6 ST part: ANY, ANYUB, NZ, NZUB, ONE and ONEUB. A general algorithm for all 6 type part is designed and we will illustrate this via an example. Consider a ST block with all ANY type parts (we use X to indicate ANY type part in the figures). We find matchings of the first part of the block. There may be many possible matchings because part of type ANY implies arbitrary matching of parts on one chain in MoDeL definitions. For each matching found, we deal with the next, and do the same thing recursively until the end of the template or matching fails. An easy example is shown in the figure to illustrate this simple idea.After all possible matchings for each chain in the template have been found, we could choose any one from each to form a combination. Matching alternatives of two different chains in the template should not be the same. For example, there are two chains in the template, T1 and T2, and three chains, C1, C2 and C3 in the species. If T1 has one matching to C1 and one to C2 and T2 has one to C2 and one to C3, the possible matchings are (T1,T2)->(C1, C2), (T1,T2)->(C1,C3) and (T1,T2)->(T2,T3). (T1,T2)->(C2,C2) is not allowed. (这里已修改)This is based on the assumption that the writer of template is always sure about the structure, he won't write a two-chain template if the parts are actually on one chain. The general matching relations could be written as (n1,n2,n3...)->(m1,m2,m3...), where (n1,n2,n3...) represents chains in the template and (m1,m2,m3...) represents the chains matched in the species for (n1,n2,n3...) respectively. However, it is only part of the matching relation because details, such as the matching positions of parts in the template, are ignored. The next question is: how to validate each matching?
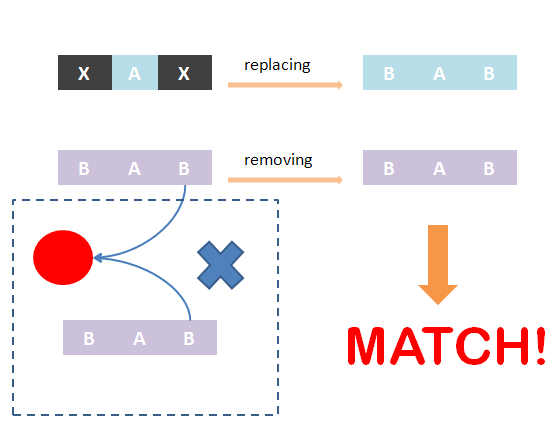
As defined in Chain-Node model, details of distribution of binding are stored in the forest. Matching of chains could not solely determine the overall match -- whether the template and the species have the same structural pattern. However, it is not easy to compare tree structure directly. We designed an algorithm to do this based on the principle: If the template and species have the same structural pattern, it must be possible to isolate the template from the species after replacing the ST part with its matchings. We first replace the ST part of the template with its matchings in the species. And then, we remove the chains which are not matched and the associated trees -- containing leaf nodes whose corresponding parts are on those chains. If the species after removing has the same Chain-Node structure with the template after replacing, the template and the species have the same structural pattern and in brief, the template could be matched to the species. We also illustrate this idea in figures.
However, there is one problem of this algorithm. Consider a LacI dimer template, whose Chain-Node model has two chains with one part on each and one tree with 2 leaf nodes. And there is also a LacI dimer species, it has the same structural pattern with the template. The matching should be (T1,T2)->(C1,C2) and (T1,T2)->(C2,C1) according to the algorithm described above. However, there is indeed one matching since the two matchings are the same. The underlying reason is that the two chains of the template are equivalent: it makes no difference to the tree structure (Huffman code of each node) if they are exchanged. Hence, several equivalent chains should be matched to a certain set of chains in the species only once.
How to create products based on database description?
In MoDeL frame, products are composed of both definite parts and ST parts. The sources of ST parts are defined in component listOfTransferTable. It seems to be very easy to generate Chain-Node structure of products from reactants and mod modifiers based on the transfer table. However, it doesn't.
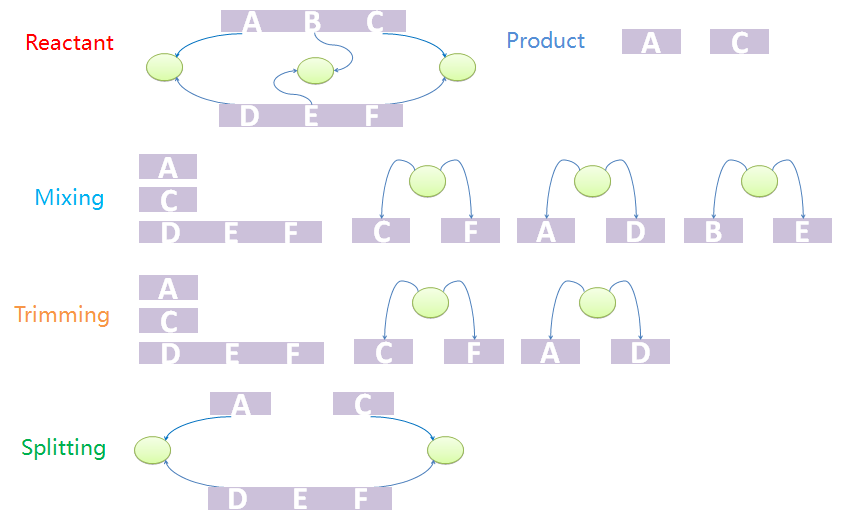
Consider first a template reaction with one reactant, A-B, where A and B are both NST parts. If it could be digested by a certain enzyme, the products should be A and B. In the real biological system, if there is a species with structure C-A-B-C, where the first part C and the last C binds to form a tree node. If we try to digest the A-B bond, the species would not be split into two pieces as expected because the two products as expected are connected by the tree node. To find products with correct structure, we design a three-step mix-trim-split algorithm to solve this problem.
The first step is to mix reactants and products. The mixture includes all chains of reactants except for those which are matched for some template chain as well as all trees. It also includes all chains and trees of products. And now we have a mixed species with chains and trees from different sources. It is worth noting that chains and trees of modifiers will not be added into the mixture. The next step is to trim the trees. Trees with leaf nodes that are not defined in chains will be removed from the forest. The last step is to split the products out from the mixture. This is because the mixture may not be one standalone complex. In other words, some chains that are not connected with others via tree nodes should not be incorporated into other species. The final standalone split species are real products in the system.
We provide an example to illustrate the product-generating procedure. The reactant has two chains, A-B-C and D-E-F. A, B, C bind with D, E, F, respectively. The products are A and C. First we mix chains and trees. Chain A-B-C will not appear in the mixture. Since node B does not appear in mixed chains, tree with nodes B and E will be removed in the trim process. Because part C and F, A and D, are both bound to nodes, the final product should contain A and B both after split processing.
In the example above, there is only one product finally though two product templates are defined in database. In this case, the corresponding reverse reaction would never be found because it fails to match two templates to only one species. This is a drawback of our MoDeL language -- it does not support description of intra-molecular reactions. If A and C could react as different parts of the product, the reverse reactions would not be lost. However, supporting intra-molecular reactions is one of our future plans for refinement of our MoDeL language.
- Resources
-
Resources
On this page we provide the information of downloading the executable files and source files of our project, and the documentation to introduce the source code build steps and system prerequisites.
Downloads
Executable files can be download at SourcForge
Project source code is available at [http://github.com/jkdirac/igame gitHub]Source Code OverView
We use [http://en.wikipedia.org/wiki/C%2B%2B C++] as our programming language, [http://www.cmake.org/ cmake] to manage our project, [http://git-scm.com/ git] to [http://en.wikipedia.org/wiki/Revision_control reversion control], [http://dinosaur.compilertools.net/ yacc and lex] to build our model file input assistant tool, [http://qt.nokia.com/ Qt4] to build the GUI, [http://sourceforge.net/projects/dbxml-core/ Oracle dbxml] as our Database manager system. Other wise, several 3-rd party open source software or tools are used in our iGame source code. [http://sbml.org/Software/libSBML Libsbml] is used to deal with sbml file operations. Qwt and Qwtplot3D are used for simulation data visual system. At the early stage of our project [http://www.tbi.univie.ac.at/~raim/odeSolver/ SBML Ode Solver] and Sundials were our main ode solver to produce the simulation data from our sbml model file, because the latest [http://www.tbi.univie.ac.at/~raim/odeSolver/ SBML Ode Solver] is 1.6.0, developed in 2005, which is out of date and fail to support the newest sbml Level 2 Version 4 standard. We succeed in updating the SBML Ode Solver 1.6.0 to support SBML L2V4, but some unfixed bugs was found, so we turn to a better ode solver [http://www.copasi.org/tiki-view_articles.php copasi], which moreover can flexibly display the ode solver output data and modify parameter to adjust the simulation output data curve. Obviously, those open source library and tools convenient to our design and implementation and help us to focus on our core working.
Build instruction
Prerequisites
To sum up, iGame should be complied and run in computers with following tools and libraries:
- C++ development environment. In Linux should be GNU C++ Tool Chain And in windows MingW Tool Chain.
- Qt4 develop and run-time environment. Qt4.7 is requirement. Editor or IDE like QtCreator or Vim is also needed.
- CMake, to configure the project and generate the Makefile. CMake 2.8 or later version is a requirement
- dbxml, The Berkeley xml native database, is available at [http://sourceforge.net/projects/dbxml-core/ http://sourceforge.net/projects/dbxml-core/]
- copasi, copasi 4.6 is a requirement. the source code and executable binary file can be downloaded at [http://www.copasi.org/tiki-index.php?page=download http://www.copasi.org/tiki-index.php?page=download], and one can follow [http://www.copasi.org/tiki-index.php?page_ref_id=115 those instruction] to build it from source code. What should be noticed is that copasi also has prerequisites, including LAPACK/CLAPACK for Maths, Qwt and Qwtplot3D for plotting, expat to parser xml, raptor The latest version of this RDF parsing library, and libsbml for deal with SBML operations.
- libsbml, Version 3.4.1 is required. Not only it is the dependence of copasi, but also the dependence of igame core code.
- Bison & Flex
- git, to download our source code by command our source code can be downloaded in [http://github.com/jkdirac/igame github], git is helpful to download.
- git clone git://github.com/jkdirac/igame.git
Build
We use cmake to manage the build progress, cmake can generate cross-platform Makefile, we intend to make our project could be built in different operating system. Until now, only Linux operating system is tested. Windows and Mac should be supported soon. Download all source code, run cmake-gui to configure the project. Pay attention to the configure information.There should be "missing ***" messages when Cmake fail to find the dependent libraries. To fix those messages, one need to assign the path of related libraries by Cmake GUI. When Makefiles are successfully generated, execute the Make command to build.
- Overview
-
One-Minute Introduction
One-Minute Introduction to
Chain-Node Model -> Template Modeling -> Automatic Modeling Database Language
Chain-Node Model
Template Modeling
Automatic Modeling Database Language
- Features
-
Features
Chain-Node Model
In most manual modeling, complex is treated as a whole entity in reactions. Plenty of synthetic biology automatic modeling software treats complex the same way. However, there is a big difference between manual and automatic modeling, that is: Model constructors always know which detail of complex needs to be ignored or preserved, but software does not. So in most software, detailed structure has to be specified by modelers, otherwise will be ignored. In most cases, this requires plenty of manual operations and thus making the software not so automatic. Knowing this, MoDeL chooses to preserve detailed structure in complex and present Chain-Node Model, which view complex as a construction of its parts throughout the modeling process. With chain and nodes, describing large biological complex with a great number of basic units and binding sites becomes possible. It also offers great flexibility in modeling. Detailed concepts are explained below.
Abstract Concept of Chain
When it comes to biobrick assembly, DNA of plasmid is often considered as an abstract straight line where biobricks can be placed on and connected together. It actually acts as the basic frame of biobrick assembly. Inspired by this, we present the abstract concept of chain, which also acts as the basic frame of part assembly. Chain is an abstract straight line where different parts can be placed on and connected together. Part is the basic unit of chain, and chain can have as many parts as possible. To go beyond DNA, and to unify the data structure of Species, we extend the concept of chain in MoDeL to include all the parts we can describe, including RNA, protein, and even compartment, compound and substituent, etc. Parts of DNA, RNA and protein, are all originated from DNA, and they are considered as “Sequence”. Sequence can be connected to other parts of sequence on a chain. On the contrary, parts of compartment and compound are considered as “Non-sequence”. Parts of non-sequence cannot be connected to any other part on a chain, which means, they can only exist on a One-Part-Chain. Substituent is used as a substitution of any other part, so it can be either sequence or non-sequence. It is important here to mention that the Sequence/Non-sequence division is only a rule guide the modeling. It is not a constraint to Chain-Node model. Actually, we choose to maximally preserve the flexibility and leave all the freedom of constructing a species to users. Users are allowed to construct ANY species they can using Chain-Node Model. For example, one can create a strange species with compartment, compound, and DNA parts on the same chain, which makes little sense in biology. So, it is really up to the users to decide how to use Chain-Node Model in modeling.
Abstract Concept of Node
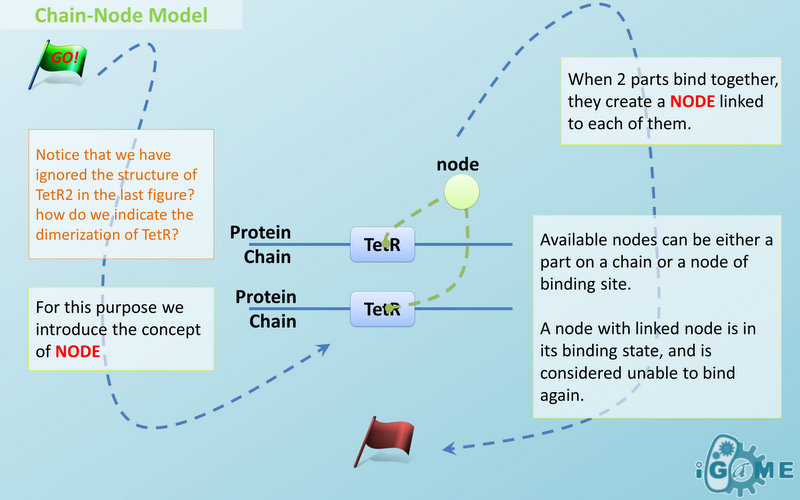
Node is used to characterize binding structure in biological complex. Node is an abstract concept of binding site or site that can be bond. Links always exist between a binding site and the sites it binds together, indicating the relationship between them. We use trees to represent the binding structure in complex. The parent node represents the binding site, and the child node represents the bond sites. For example the binding structure of TetR dimer is considered as a TetR2 node with two TetR nodes as its children. There is one exception though, the ROOT node. ROOT node is used as the top node in a tree. It can only have one child and no parent. The function of ROOT is to indicate that its child is the last binding site in this tree, and it binds to nothing any more. In the reactions of promoter repression and activation, the promoter is repressed or activated after binding with repressor or inducer. So its properties are changed, and it is not the original promoter any more. This is an example where binding changes the properties of node. Since we do not store all the possible property alterations within one part, the solution is to replace the old part with a new part that stores the correct properties. Available nodes can be both a part on a chain or a node of binding site, which leaves a lot of possibilities in constructing complicated binding structure. Again, there are no restrictions, but parts of compartments are not suggested to appear in a binding structure.
The Basic Assumption
The basic assumption of Chain-Node Model is that a part ALWAYS carries its properties wherever it is placed in an instance of Chain-Node Model. This means no matter how complicated the complex is, you can always keep track of the location and properties of every single part. By letting complex inherits its parts’ properties, the assumption saves modelers from rewriting the structure and function of each new complex. However, we should address that the assumption is not suitable for all circumstances. For example, under this assumption, a fusion protein will carry the properties of the fused proteins, which is not true in many cases. A fusion protein may not remain its old properties and even have new properties that none of the original parts have. In this case, our suggestion is to define the fusion protein as a new part. So it is suggested that users choose different strategies of describing the same thing in Chain-Node Model base on different situations, instead of modeling everything in the same way.
Examples
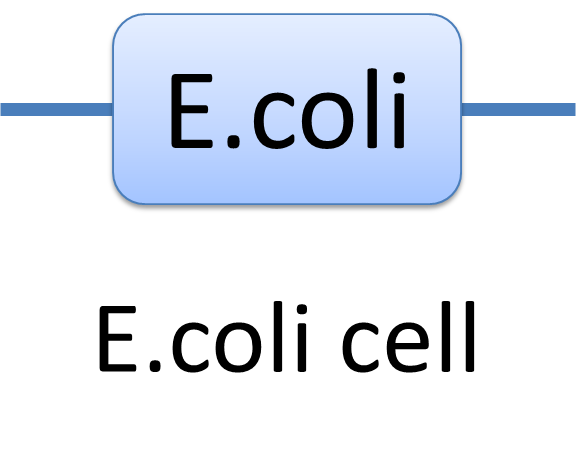
With great power there must come great responsibility. In order to use the powerful Chain-Node Model correctly, please have a look at a few examples we presents. Example of a species: E.coli cell In MoDeL, the cell of E.coli is described as a species with only one part of E.coli(see Figure 1). Since the concept of part has been extended greatly, living cells can be described in the same way other species. Compartment belongs to “Non-sequence”, so it is suggested that E.coli only exists as a one-part-species.
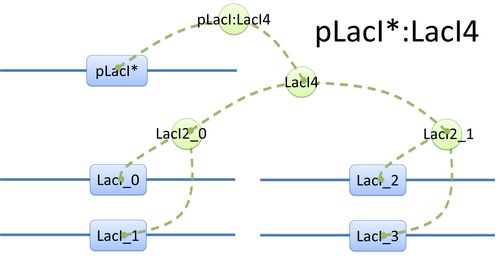
Example of a species: pLacI*:LacI4 and its tree structure of binding. Here is another example of template species with more complicated structure.(see Figure 2) In biology, LacI tetramer (or LacI4) can bind with promoter pLacI as a repressor. When bond with LacI4, pLacI is repressed and its transcription speed will decrease significantly. In order to express this alteration, as mentioned above, we use another part of pLacI* with decreased transcription speed instead of pLacI in the species.
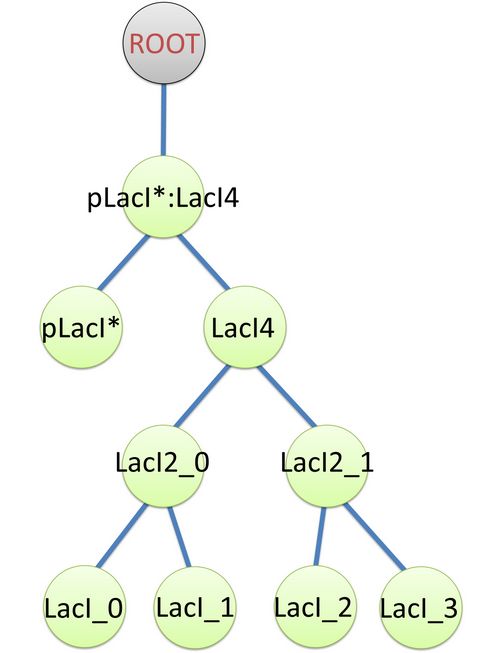
The structure of binding sites is shown in figure (see Figure 3). The graph indicates that LacI monomers first bind together to form LacI dimer, then LacI dimers forms LacI tetramer, and finally LacI4 binds with pLacI* to form pLacI*:LacI4. Modeling with Templates
Manual modeling of biological system is widely used in synthetic biology. However, it requires an overall understanding of the biological network and large amount of data, making it difficult even for professionals. Actually, the data provided manually are redundant because different reactions may occur with a same mechanism. Based on this idea, we introduce the concept of Template, which is the minimal data set for describing biological reaction networks, as a feasible way to implement automatic modeling. The term “automatic” does not mean modeling without providing any information. Instead, we aim at providing minimal amount of data. Similar to C++ programming language, templates allow generic description of species of a certain structural pattern or reactions of a certain reaction mechanism. Substituent and Template
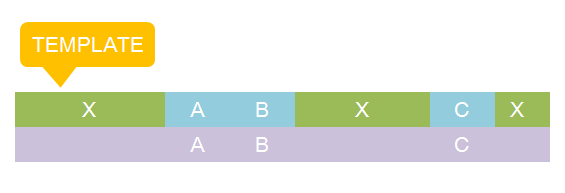
There are two kinds of templates: Template Species and Template Reactions. They are designed to represent a family of species with similar structure and a family of reactions with similar mechanism respectively. In order to realize this idea, we add the container of Substituent into MoDeL. There are currently six objects in this container, ANY, ANYUB, ONE, ONEUB, NZ and NZUB. Though defined differently, they actually serve the same purpose: representing other parts. Let’s take ANY as an example (see Figure 4). ANY, represents parts on ONE chain and the number of represented parts can range from zero to infinity. For example, a DNA species with structure ANY - pTetR - ANY represents any DNA with part pTetR. A reaction with template species as its reactants, modifiers or products is a Template Reaction. A template reaction describes the core structure of reactant that causes the reaction to happen and provides a specification for generating reactions with the same mechanism. This could be understood more clearly by dividing parts in a species into functional groups -- a reaction template describes the interaction mechanism of these functional groups. For example, pTetR promoter is deactivated in presence of TetR dimer. The template species are pTetR DNA template and TetR dimer protein template (see Figure 5) and the functional groups are pTetR promoter and TetR dimer. All the parts of ANY should also be counted as a functional group, though they don’t have functions in this reaction. Any pair of species which partially contain pTetR DNA and TetR protein dimer respectively would bind according to the description of this reaction template. Modeling with templates allows users to define species and reactions only once for one certain family without rewriting them again in database. We are still looking for more substituents. We are planning to have more detailed substituents that can replace certain number of parts on certain number of chains. Even the arrangement of parts and chains can be specified. We believe it will make our template modeling more powerful.
A Standard Language
MoDeL aims at providing a language and syntax standard for automatic modeling databases used in synthetic biology models. We will discuss a few relevant topics in the following sections. Standard Biological Part Automatic Modeling Database Language
With plenty of information for automatic modeling needs to be stored, we design our database, and develop a standard language of describing the database. We call the language: MoDeL. MoDeL (Standard Biological Part Automatic Modeling Database Language) is a database representation format for synthetic biology automatic modeling. This project of MoDeL aims at providing a language and syntax standard for automatic modeling databases used in synthetic biology models on a number of topics, including cell signaling pathways, metabolic pathways, biochemical reactions, gene regulations, and so on. MoDeL is developed as a machine-readable format in XML fashion, programming languages with XML library API (such as libxml2 for C++) are strongly recommended for reading MoDeL database. USTC_Software has been devoted to the standardization of MoDeL. Documents can be found here. Example of database written in MoDeL can be downloaded on our [http://github.com/jkdirac/igame GitHub page]. MoDeL is an attempt to define a universal database markup language for synthetic biology automatic modeling. It supports descriptions of a wide variety of different parts, including biobrick, compound, compartment, substituent, and so on. It can also describe the species constructed by these parts, and reactions happen with the species, such as binding of species, dilution of species, degradation of proteins, amplification of cells, multi-compartment reactions, and so on. Many traditional methods fail to consider structure of species when trying to markup reactions with only reactants, modifiers, products and kinetic laws. The best part of MoDeL is that it can preserve detailed structure in species and reactions using Chain-Node Model. Based on the model, MoDeL is able to markup almost all kinds of reactions in a detailed and clear way. Overview of MoDeL
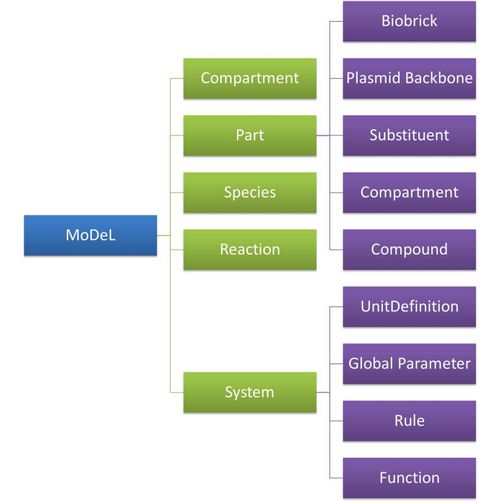
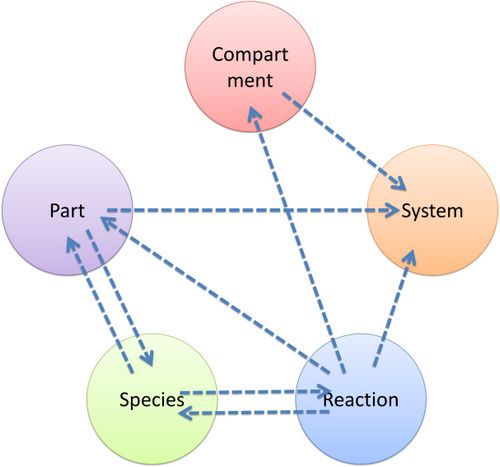
Figure 5 above is an example of a simple network of biochemical reactions(pTetR + TetR2 = pTetR*:TetR2) that can be automatically constructed using MoDeL: MoDeL supports automatic construction of such network with only the input of its initial conditions and environmental parameters. In order to do this, obviously the data of parts, species, reactions, compartment, units and so on is needed. Besides, relationships between these components are also needed, so we can link to a reaction when we find a species. Data of relationship is stored as references and exists in most MoDeL components. To define all the needed components in a systematic way, MoDeL organizes them in five component containers at the top level: :: System: This container contains definitions of units, functions, rules and global parameters. These are all basic elements for other components of MoDeL, and many are mathematical concepts without direct synthetic biology meanings. The name of the container, System, separates this container from others which contains components of biological significance, and indicates this container includes the basic parameters of the whole system. :: Compartment: This container contains definitions of compartments. Compartment here is considered as a holder of substance with certain volume, so it only stores related data such as units and volume. It also stores a list of compartments that are permitted to be placed inside. For compartments like E.coli that can multiply, a corresponding compartment with the same id will be created in the sub-container of compartment in Part. :: Part: This container contains definitions of parts. Part includes the basic units that can be inserted in an abstract chain. The container of Part has five sub-containers which are: compartment, biobrick, compound, plasmidBackbone, and substituent. Biobrick contains biobricks from PartsRegistry together with other similar components created by users. Compartment contains cells and other microscopic organisms that can multiply. compound contains small molecules like IPTG and other compounds. PlasmidBackbone contains plasmid backbones come from PartsRegistry or defined by users. Substituent contains substitutes for template modeling. :: Species: This container contains definitions of various species defined by Chain-Node Model. Each component stores the cnModel structure of chains and trees. Related reactions are also stored. It is suggested that the constructor of a database stores template species instead of real species in this container. Only in this way can he make use of the power feature of template modeling of MoDeL. :: Reactions: This container contains definitions of various reactions. Each component has its own compartments, reactants, modifiers, products, substituent transfers, and kinetic law. This forms a detailed description of reactions. Again, constructor of a database is suggested to store template reactions instead of real reactions. The organizational structure of components in MoDeL is shown in the following figure. As mentioned above, reference relationships are stored in most components. The following figure shows the relationship between different components, the pointer pointing to a component indicates referencing to that component. It is worth noting that MoDeL does not attempt to include all information about each component. In PartsRegistry, a long list of data in XML format is provided including part author, status, sequence, and even group access information. Such data are redundant for our database since they are not necessary elements needed to construct reaction network. Only the useful data is retained. Relationship with SBML
SBML(The Systems Biology Markup Language) is a machine-readable language in XML fashion. It's oriented towards describing systems where biological entities are involved in, and modified by, processes that occur over time. As a widely used model exchange standard, SBML is chosen to be the output of our auto-modeling module. Generally speaking, MoDeL needs not to be conformed to any constraints set by SBML. However, since SBML is a widely used standard, it is better for MoDeL to share common concepts and components with SBML as many as possible. For example, MoDeL still uses rules, function definitions and unit definitions in SBML. By doing so, it will be easier for software tools to change formats between MoDeL and SBML. - Language Specifications
-
Language specifications
History (optional)
The simplest example with only E.coli cell
introduction
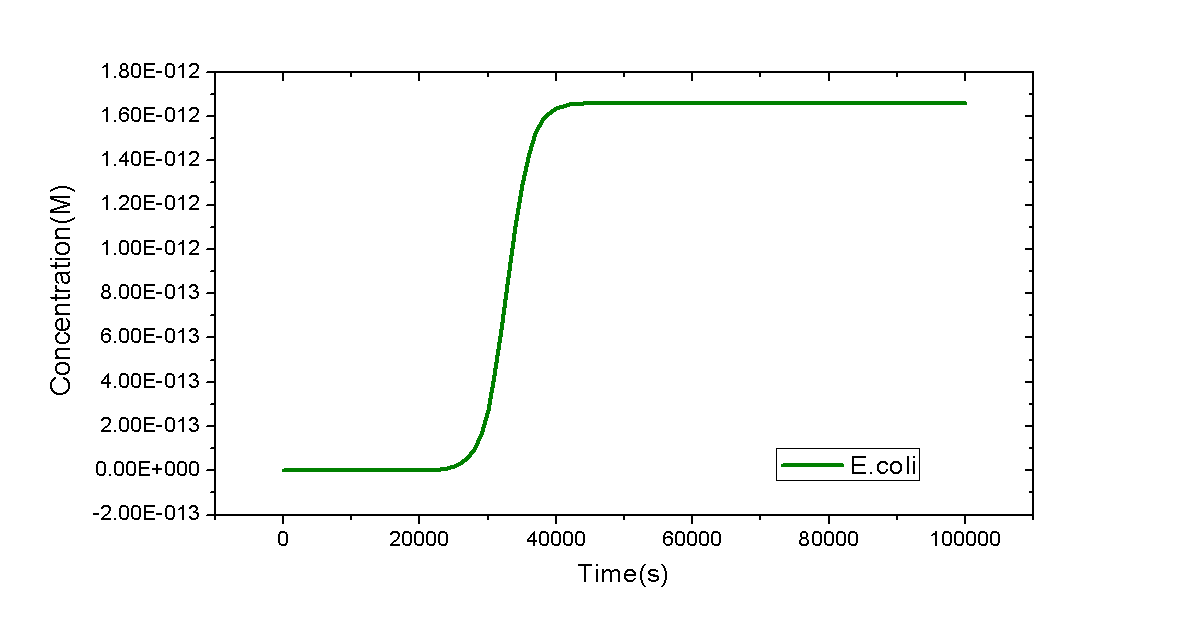
As the simplest example, a model of a flask with only E.coli cells inside will be shown. Only one reaction will be considered: the replication of E.coli. However, it is not trivial. MoDeL enables users to add such an auto-catalytic replication reaction easily and conveniently. The minimal database used for this model could be download here.
database construction
We only provide key points regarded with construction of this minimal database. To add the auto-catalytic reaction in the Reaction container, a species with only part E.coli is required. It has the simplest Chain-Node model format: only one chain with one part and no trees. The auto-catalytic reaction has one modifier and one product and they are both referred to E.coli defined in Species container. Since they are compartment-type species (species representing a compartment), attribute itself of compartmentLabel node in modifiers and products definitions should be set the same with label of the compartment they represent in the compartments definition. It ensures that the product and the modifier are the same, avoiding wrong mismatch of the product which is different with the modifier. Since number of E.coli cells will reach a stable level in a long time course, we use <math>k_{g}(1-C_{E.coli}/C_{max})C_{E.coli}V_{Flask}</math> as the reaction rate, where <math>k_{g}</math> is the growth rate of E.coli, <math>V_{Flask}</math> represents the size of E.coli, and <math>C_{E.coli}</math> and <math> C_{max}</math> are concentration of E.coli and its max concentration in the flask, respectively. The negative sign in the rate equation indicates self-repression of E.coli cells.
modelling and simulation
Model written in SBML format is shown below. The dynamic curve of E.coli replication is plotted by time course simulation. Exactly speaking, dynamic behaviors of biological system are meaningless after 4000s because of termination of reproduction of E.coli. However, the curve is in qualitative agreement with that from experiments in the first and second stage of reproduction of E.coli.
Basic example with pLac-LacI repression system
introduction
MoDeL provides poweful supports for description of interactions between two species. No interactions exist in the previous example. In this example, we will construct a plasmid backbone with promoter, rbs, coding sequence and terminator inserted in and transform it into E.coli. LacI repressor are produced after two steps of transcription and translation. It binds with itself to form a dimer and then a tetramer, which could bind with promoter LacI on the plasmid to repress the process of transcription. Besides, replication of E.coli cells, degradation of proteins and mRNAs, as well as dilution due to replication of cells, are also considered.
database construction
Actually, it is a complex molecular process and takes many steps to complete a transcription reaction. It includes binding and initiation, elongation, and termination. So does translation reactions. The first version of MoDeL DOES NOT support transcription and translation reaction templates and they will be handled by our core program of iGame and thus no need to write in the database.
All binding reactions in the database demo are modeled using law of mass action. They are dimerization of two LacI repressors and that of two LacI dimers. LacI tetramers will bind with promoter LacI in the plasmid and thus repressing the expression of plasmid DNAs. However, MoDeL enables any mathematical model form for species-species interactions and is not constrained within only law of mass action. Users could change the reactants, modifiers, products, as well as kinetic laws arbituarily and validate their models via output of dynamic curves.
Species reaction templates are worth mentioning to show features of our new language MoDeL. It is never a easy problem to calculate degradation rates of proteins since different parts in a fused protein may have different degradation rates and the overall rate is still unknown. What's more, a protein in MoDeL may have different protein chains and it is more complex to determine the overall rate. In the demo database, we assume all proteins with no parts in bound state have a uniform degradation rate. Proteins with more than one chains are considered stable and non-degradable. This idea could be applied easily by writing reaction templates in MoDeL format. The reactant is a general species template with only one substituent-type part (ANYUB) of type ForwardProtein (no reverse reactions happened here). It is similar for degradation of mRNA molecules. Only mRNAs with single chain degrades at the same rate.
For reactions of dilution due to reproduction of cells, quantities of species decrease in a similar way to degradation. The rate of dilution is also similar to the reproduction rate of E.coli:
<math>k_{g}(1-C_{E.coli}/C_{max})C_{species}V_{Flask}</math>
where we only replace the second <math>C_{E.coli}</math> with concentration of species in dilution. To apply this idea, we only need to construct a species with only one substituent-type part (ANY) of no specific type and it will be matched to all species. Among them, only species bound in compartment E.coli will be diluted.
Replication of plasmid DNAs is not as easy as reproduction of living cells in two aspects: first, plasmid backbone with the same replication origin could be found in many species in the system because the original transformed plasmid backbone may bind with other compounds or proteins to form complex; second, the replication of original transformed plasmid DNA molecule tends to be repressed by all species including the same replication origin. However, E.coli cells do not have any effects on the reproduction of other E.coli cells. MoDeL language perfectly support this idea by separating the template reaction into replications and repressions. The first part describes the exponential amplification of plasmids with reaction rate <math>k_{g}C_{plasmid}V_{E.coli}</math>, where <math>k_{g}</math> is the replication rate of a certain plasmid, <math>C_{plasmid}</math> is the concentration of this plasmid, and <math>V_{E.coli}</math> is the volume of E.coli cell. The product may be different from the reactant. It is possible because the reactant may be a complex, such as complex of LacI tetramer and plasmid with promoter LacI (we call it pLacI:LacI4 below). Only the products of the replication reaction will be repressed with a certain rate. In this example, pLacI:LacI4 would not be repressed since it is not among the products of its replication. Via MoDeL language, we design a template reaction with no reactants, two modifiers and one product. The two modifiers are template species containing a certain plasmid backbone (say pSB1A3), and binding state is allowed for both. The product is also a template species containing pSB1A3, however, it has only one chain and no binding is permitted. Hence, modifiers have Chain-Node structure ANY--pSB1A3--ANY, while product has its structure ANYUB--pSB1A3--ANYUB. The listOfTransferTable makes parts transfer from part ANY of either modifier to its corresponding part ANYUB of the product. The reaction rate is <math>-k_{g}C_{mod1}C_{mod2}/C_{max}</math>, where <math>k_{g}</math> is the same as that defined in the replication reaction, <math>C_{max}</math> is the max concentration of plasmids containing pSB1A3 as backbone, and <math>C_{mod1}</math>, <math>C_{mod2}</math> are concentrations of modifier1 and modifier2, respectively. The negative sign in the rate law indicates a repressed effect to the replication of the product.
modelling and simulation
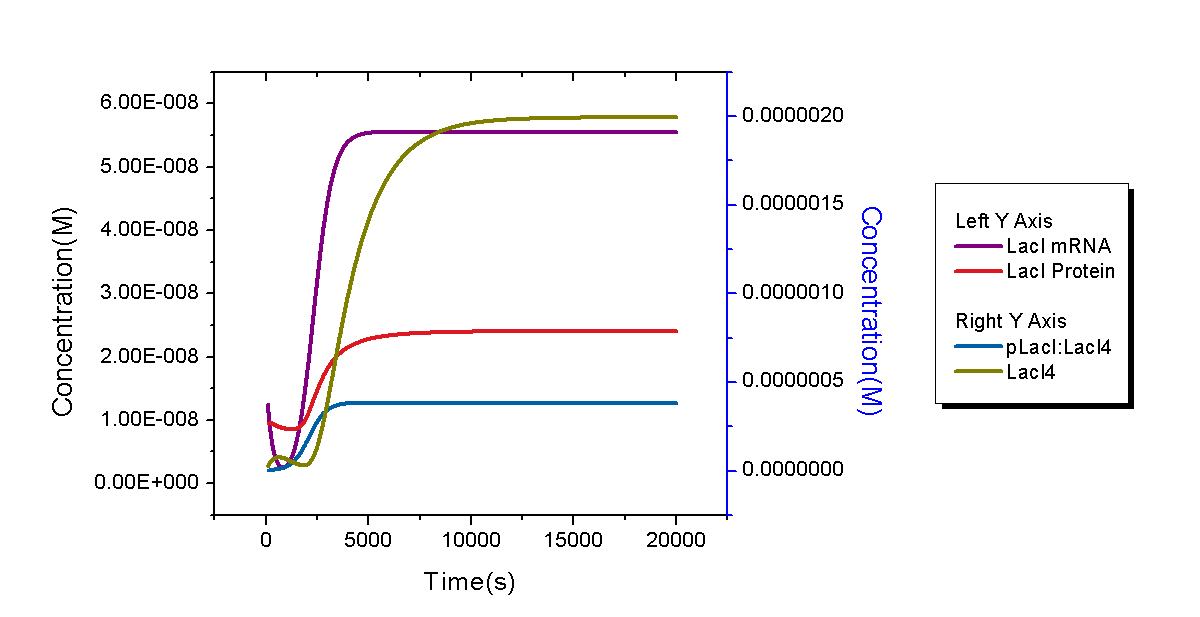
Only one copy of plasmid is transformed into E.coli cell at initial time. To give a reasonable dynamic curve of species in the biological system, parameters are obtained from experiments if they are available. Besides E.coli, there are 6 species: DNA with pLacI and LacI (DNA0), LacI mRNA (with RBS), LacI protein, LacI dimer, LacI tetramer, and complex of LacI tetramer and DNA0 (DNA1). Concentrations of LacI mRNA, protein, dimer and tetramer versus time are plotted in Figure 4. Since dissociation constant (Kd) of LacI-LacI monomer interaction (77nM) is mush larger than that of LacI dimer-dimer interaction (0.1pM), steady-state concentrations of LacI mRNA and protein are in different scales with that of LacI dimer and tetramer. Due to very large Kd of LacI dimer-dimer interaction, LacI protein would finally exist in form of tetramer in the system (concentration of LacI dimer is several orders of magnitude lower than the scale of the left Y axis and thus be ignored).
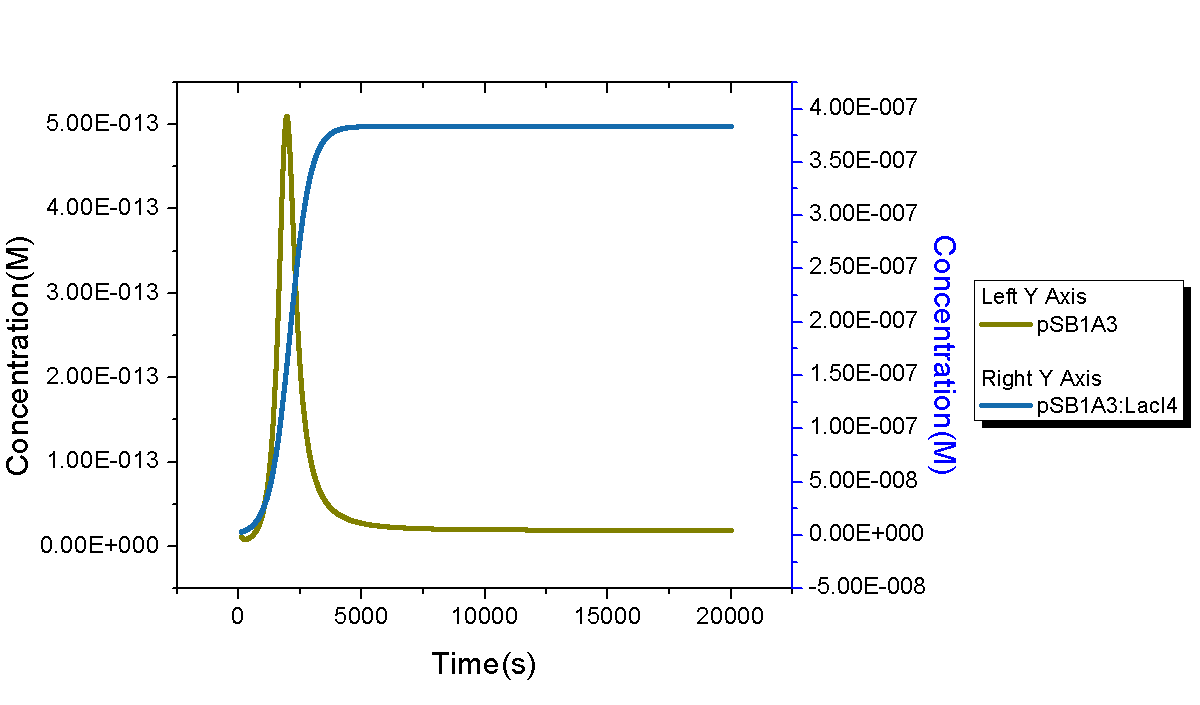
Figure 5 shows dynamic behaviors of plasmids in our model. Since only one copy of plasmid DNA0 was transformed, its quantity increases at the very beginning and decrease soon because all DNA0 will bind with LacI tetramer and no free plasmid exists. The process will end in one hour and quantities of both DNA0 and DNA1 enter into a stable level. It is worth mentioning that the stable concentration of DNA1 is about 380 nM, a little lower than its maximum concentration 475 nM. It is owing to reproduction of E.coli cells. After quantities of E.coli cell go into its stable stage, its concentration will continue to increase until the maximum setup value.
This basic example proves great success for iGAME to automatically model genetic regulatory systems. It shows real biological process in a qualitative manner and give suggestions for users to adjust their experiment plans.
(# ̄▽ ̄#)
A big step: IPTG-induced pLac-LacI system
Demo with reverse parts(Demo of toggle switch)
Demo with environmental conditions
Demo with substituent transfers
Demo with multi-compartment reactions
==Demo with complex binding product==
Lab Notebook
Meetings are set every Saturday since Feb.15th, then three more discussions (Mon, Wes, Fri) are added when we start to determine our project since Mar 21th.
- 2010.4.10 – 2010.5.14 reports of the former iGEM works
- 2010.5.21 – 2010.6.11 broach of candidate projects
- cell density control
- Reporter and imaging
- DNA writing and reading in the cell
- Biodegradation of plastics
- 2010.6.12 – 2010.6.30 further research by small groups on
the candidate projects
- sketch map of our final project host-cell free reaction
chamber was set up
Month NoteBook
| March | April | May | June |
| July | August | September | October |
Dialy NoteBook
|
|
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
However, we do not stop here. We paid great effort to the safety issue and we have been dedicated to improving the knowledge of synthesis biology from classes and tried to share the part we contribute to the worldwide. We innovatively plot a new way of spreading the spirits of IGEM by starting our own community and we will never stop sharing and communicating.
- Overview
-
Team Overview
This year 10 students started the iGEM USTC Software team: Chen Liao, Zheng Wang, Kun Jiang, Jue Wang, Luojun Wang, Yubo Qi, Zhaoyi Li, Zhengda He, Shangyu Luo and Mort Yao. There are only 8 of us in the picture, while the only female team member, Jue Wang was in Beijing engaging for her TOEFL exam, and Shangyu Luo had gone to Hong Kong for studying in The University of Hong Kong as an exchange student. What a pity! But they will join us at the iGEM 2010 Jamboree.
USTC has formed two IGEM teams this year, one wet lab team and one dry lab team. As many students majoring in biology was attracted to the wet lab team, our team consists of students from other departments, such as Mathematics, Physics, Computer Science and so on. Thanks to our instructors Hao Jiang and Zhaofeng Luo, with their professional instruction, we are guided to decide the project plan, design and implementation.
About iGEM@USTC
The iGEM Team of USTC are the combination of the exact tradition of USTC and the exact vitality of the young. Rigorous daily schoolwork has provided the team members with a solid foundation of basic knowledge, and an innovative spirit enables us to stand on the frontier of the modern science.
In 2007, Jian Zhan, Bo Ding, Rui Ma, Xiaoyu Ma, Yun Zhao, Ziqing Liu and Xiaofeng Su, guided by Prof. Haiyan Liu, Prof. Jiarui Wu and Prof. Zhonghuai Hou, won the "Best Foundational Technology Prize" and a Gold Medal, with the project "Extensible Logic Circuit in Bacteria".
In 2008, Xunyao Wu, He Shen, Genxin Liu, Jia Liu, Binjie Xu, Yanting Xue, Yu Chen, Zhen Xu, Bo Ding and Jian Zhan, guided by Prof. Haiyan Liu, won a Silver Medal, with the project "Artificial Multi-cellular Self-organization System".
In 2009, Hao Jiang, Li Xing, Hao Wu, Zongxiao He, Danqian Liu, Chao Li, Jiayi Dou, Hanyu Lu, Hao Zhou and Bing Wu, guided by Zhaofeng Luo, Jian Zhan, Jiong Hong and Xiaoxiao Ma, won a Gold Medal, with the project "E. coli Automatic Directed Evolution Machine (E.ADEM)".
In 2009 we also formed a software team for the first time. Wei Pan, Yuwei Cui, Jiahao Li, Yu He and Xiaomo Yao, guided by Bo Ding, also won a Gold Medal, with the Project "Automatic Biological Circuit Design (ABCD)".
Since last year we have accumulated some experience from the project of Automatic Biological Circuit Design (ABCD), we expect to develop a synthetic biology game this time. The game aims at teaching non-biologists how to design biological systems so that players can compete to improve their designs in order to help biologists create the best engineered machines!
For more information, go to [http://biotech.ustc.edu.cn/forum/forumdisplay.php?fid=26 USTC iGEM Forum].
About USTC
- Message from the President
A university is the important assurance of maintaining belief, developing science and culture. It is a significant symbol of the level of civilization achieved by a nation.
- USTC Today
The University of Science and Technology of China is a new type of university established by the Chinese Academy of Sciences (CAS) after the founding of People's Republic of China. The establishment of the University is aimed at fostering high-level personnel of science and technology absolutely necessary for the development of the national economy, national defense construction, and education in science and technology. The University was founded in Beijing in September of 1958. Mr. Guo Moruo is the first president of the University...
- History of USTC
"Pooling the entire strength of the CAS for the development of USTC and combining the institutes of the CAS with the departments at USTC" "Studying hard and excelling in both integrity and specialty"USTC was established in Beijing in September, 1958 under the presidential leadership of Guo Moruo. The establishment was hailed as "a major event in the history of Chinese science and education"...
For more information, please visit [http://en.ustc.edu.cn/ http://en.ustc.edu.cn/] (in English) and [http://www.ustc.edu.cn/ http://www.ustc.edu.cn/] (in Chinese).
Team Cooperation
iGEM 2010 China Meetup Shanghai 5/6th Aug, 2010
On August 5th, teams from 7 top universities in China, SJTU-BioX-Shanghai, ECUST-Shanghai, Peking, Tsinghua, USTC & USTC_Software, ZJU-China, and Sun Yat-sen University all gathered in Shanghai and had a nice day at the iGEM 2010 China Meetup.
The meetup started around 9 with a short speech by Professor Lin He, head of Shanghai Jiao Tong University Bio-X Institute, which hosted the event. Then each team made a presentation displaying their projects and progresses since the summer began, followed by free questioning and discussion time. And when it was near the end of the day, student members had a hour or two to talk and play freely with each other, with a big meal waiting ahead for them.
For more information, go to https://2010.igem.org/Events/China_meetup.Gallery
- Students
-
Student Members
I am soumort! I have nothing to say!!
My Profile:
Chen Liao, Room 418, Key Lab of Quantum Information, CAS, East Campus, USTC Hefei, Anhui 230026, China, liaochen@mail.ustc.edu.cn
I’ve just began my first year in the Optics and Optical Engineering program as a master candidate at University of Science and Technology of China. I became interested in iGEM because I found it a great chance to learn synthetic biology. Since iGEM is a student-run and student-manage programe which encourages students both to work collectively in a team and work individually to complete one's own task, it is a great experience to exercise my team working ability as well as creative ideas. In the last months, especially the summer, I worked with students from different disciplines to design and implement our project together and have learnt a lot from them. It is rather a wonderful experience to work with all team members. (^▽^)
As a student of department of physics, my research area is development of first principle theory and implementation of LCAO (Linear Combination of Atomic Orbitals) algorithms. As a member of [http://lqcc.ustc.edu.cn/helx/ LiXin He's group], I have successfully implemented atomic force calculation in LCAO method and my next project is to modify and develop algorithms under condition of magnetic B field. Besides scientific research, I do sports very often. Basketball is my favorite and my favorite basketball player is Kevin Garnett (K.G.), his professional attitude on the court as well as his individual character in the life impresses me a lot.
I am Zhen Wang!
Zhen Wang, Room 418, Key Lab of Quantum Information, CAS, East Campus, USTC Hefei, Anhui 230026, China. Email: xqqixp@mail.ustc.edu.cn; wangzhen.ustc@gmail.com;
Hi, I'm Zhen Wang, I mainly work on MoDeL and Database of iGAME. As a member of Lixin He's research group, I focus mainly on calculation of quantum information, quantum correlation and entanglement.
You might have noticed that my address is same as Liao's. Yes, we are from the same Research Group, and the same department, too. It will be boring if I repeat all the information again, so I am going to write some experience of myself participating iGEM2010.
As a student majoring physics, it is hard for me to believe that I can participate in a biology project. Luckily, to my great surprise, these two areas are not so far apart. Modeling and analyzing skills of physics can also be used in biology, and dry project always means a lot of coding task, I can handle that, too. Of course, there is plenty of biology I need to learn, but with the help of our instructor and wet team members, I learn it fast. So I want to tell everyone that there's no limit in iGEM, join in and unleash your power!
Email: jkdirac@mail.ustc.edu.cn
I am Kun Jiang, the cheif software developer in iGem 2010 USTC_Software team, responsible for designing the global architecture of our software and developing the ODE Solver module, Berkley database interface module and several simple tools for teammates.
It is my second year of studying in computer applied technology lab of Institute of Plasma Physics Chinese Academy of Sciences as a master candidate. I am interesting in software engineering and Computer Sicence. Last year I noticed the recruit advertisement of iGem 2010 USTC_Software team in school BBS website. I make the decision to apply immediately. It's really happy for me during those past server months working with our team.
I play Ping Pong most during my spare time, because Ping Pong is a sport emphasizes on speed, skill and acting harmony ability rather than brute force. And I go jogging every day, which i think is a good way to relieve pressure and keep me in health!
I want to be a brilliant software engineer developing some brilliant works. I gain strong knowledge in software engineering and computer algorithms. IGEM 2010 is one step to my ambitions, it is a important project but not the last project of my engineering life, I wish I would develop more and more softwares in future. If you need help, do not hesitate to contact me please!
I am Yubo Qi!
Yubo Qi 3005, national labotarory for physical sciences at the microscape
I am an undergraduate student in theoretical physics. But I am also interested in chemical physics and biology. In my labotarory, I concentrate on the calculation about transmission in nanostructure. However, biology science is also my favourite. So, I decide to join USTC_Software team of iGem 2010. Now I write database of biology reaction for my team. I love biology, chemistry, physics and decide to devote myself to the research of interdisipline in the future.
Luojun Wang(Vic Wong)
Dept. of Statistics & Finance, USTC, Hefei, Anhui 230026, China;
Lab of Systems Biology of Hefei National Laboratory for Physical Sciences at the Microscale, USTC, Hefei, Anhui 230026, China
vicwong@mail.ustc.edu.cn; victorwangluo@gmail.com;
Hey, I'm Vic, an undergraduate senior student majoring in mathematical statistics. Yet my research field is biology statistics, and that is why I'm here.
For IGEM, I'm mainly responsible for human practice and have been giving lectures as well as other promotions on both synthesis biology and IGAM. It is this special experience that has not only made my college life amazing, but also presented me with a broader view of biology. Besides working for IGEM, I've also been researching biology statistics, including bioinformatics and biology networks.
My goal is to be a biostatistician in the future, on which I'm gonna keep putting effort. Now I'm applying for an opportunity to study as a biostatistics PhD. Good luck to me, huh? : )
I am Jue Wang!:)
Dept. of Math, USTC, Hefei, Anhui 230026, China;
Email: juewang@mail.ustc.edu.cn
Hi,I am Jue, a Math majored senior student. As the boundaries among different disciplines of science are now blurring, I want to handle the ideology of maths to arrive at a higher standpoint to try to understand the principle of life. iGEM provides us with such a good state to learn from utilization.
In this group, I am the unique girl among nine boys, and the most frequently spoken of mine is "Oh, it's my duty to get some soy sauce! " Strange, huh? The connotation of it is not words but meanings. Those guys are too gentle to arrange me doing something of coding.So I am more involved in clerical works, I design the team uniform and our logo,arrange the scheduling and get in touch with sponsors and alumni connection. My life is full of unpredictable tasks, who can tell me what is the next?
My profile:
Zhaoyi Li, Hefei National Laboratory for Physical Sciences at Microscale & National Synchrotron Radiation Laboratory, East Campus, USTC Hefei, Anhui 230026, China, lizhaoyi@mail.ustc.edu.cn </p>During my 2 years’ journey in the nano world, I got to know iGEM and joined in USTC Software team with great honor. Just as NEW YORK TIMES says, iGEM is a Do-It-Yourself Genetic Engineering Program. I am strongly attracted by this idea and cannot help imaging the science fiction picture where our daily life is changed by our genetic machine. It is unforgettable to have a brainstorm with talents from different academic background and turn our imagines into reality. Thanks to iGEM, I experienced a new trip in the synthetic biology and benefited a lot from my partners. Also, to compete against more than 1,000 other students from 100 schools, including many top-flight institutions is exciting.
I love traveling and always enjoy trips with my friends. I have a dream that one day I can travel around the world to discover the great scenery make new friends and experience different culture. Sports is also my favorite, I am good at playing football and gifted in table tennis.
I am Zhenda he!
Hello , everyone! My name is He Zhengda , a student from Chemical Physics Department in University of Science and Technology of China . To be a member in 2010 USTC_Software is a really great time for me during the boring semester ! I met a lot of interesting people in the IGEM lab , and I wish to meet all you guys during the Jamboree . I have a enormorous interest in math , computer science and designing stuff . I am a Wiki interface designer in our team . So I hope everyone like it . It's a short description of me~ See you in the Jamboree!
I am Shangyu Luo!
Introduction: Hi all, I am Shangyu Luo. It is my Chinese name and my English name is Seraph. I am very pleased with attending iGEM competition with my teammates of USTC SOFTWARE. My major is computer science and I am interested in both computer and biology. During this competition, I gain a lot and make many friends. Thanks for my teammates, teachers and School of Computer Science and Technology! Additionally, I favor AI (Artificial Intelligence) in the fields of computer science, if you would like to create some beings together with me and to make them intelligent, please feel free to contact with me! Thanks all!
By Seragh
lsyurd@gmail.com
School of Computer Science and Technology
University of Science and Technology of China
- Instructors
 "
"