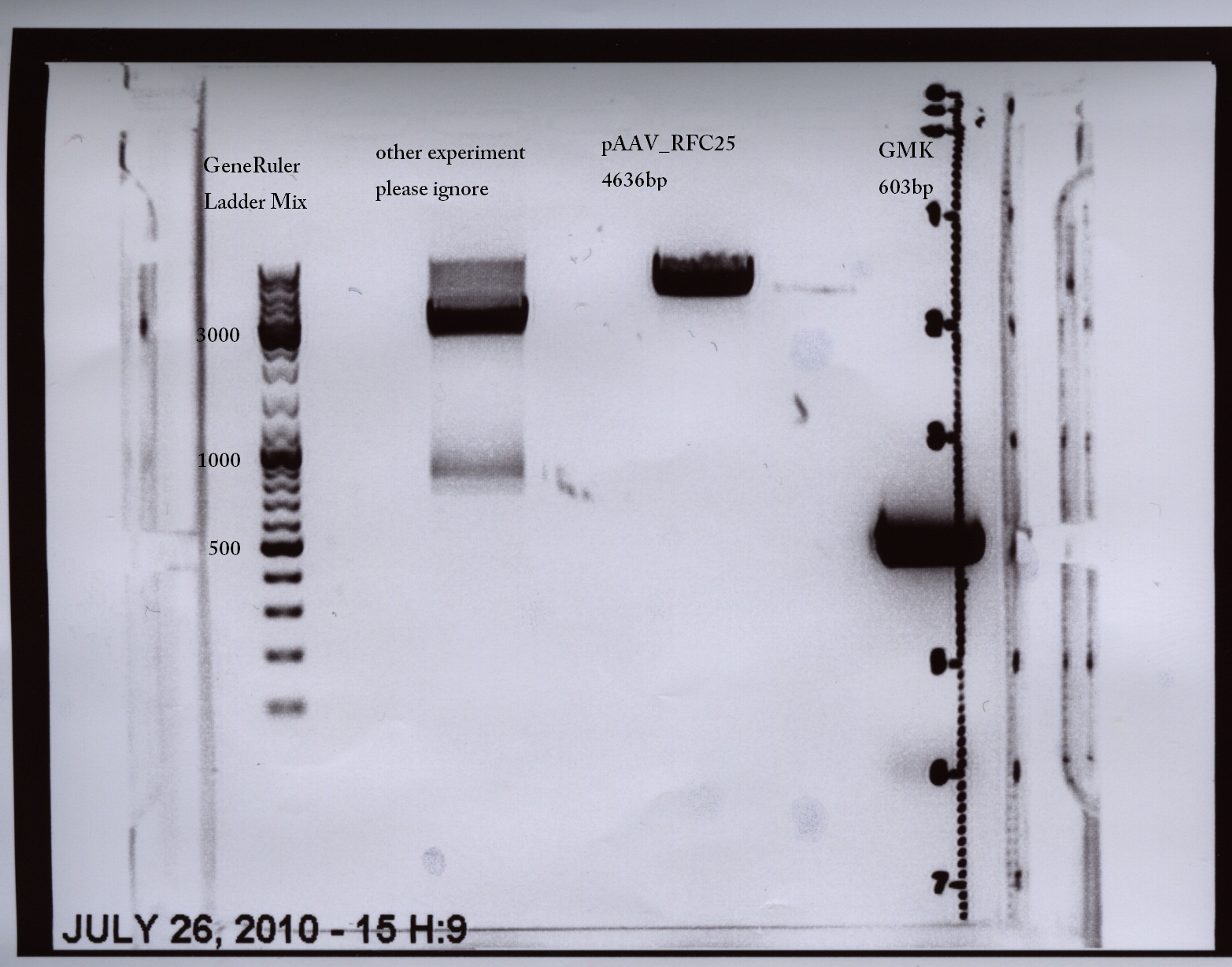
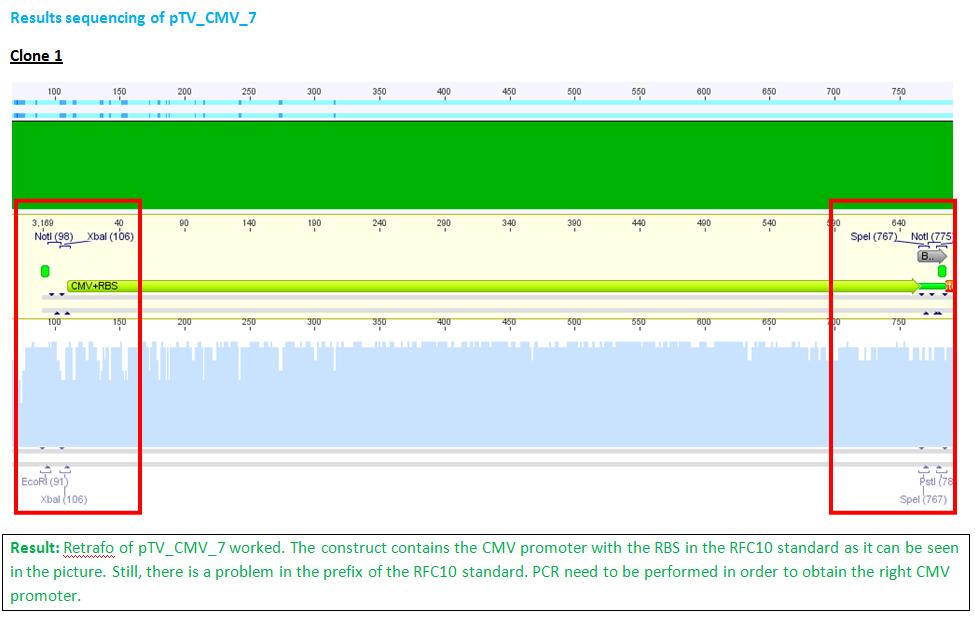
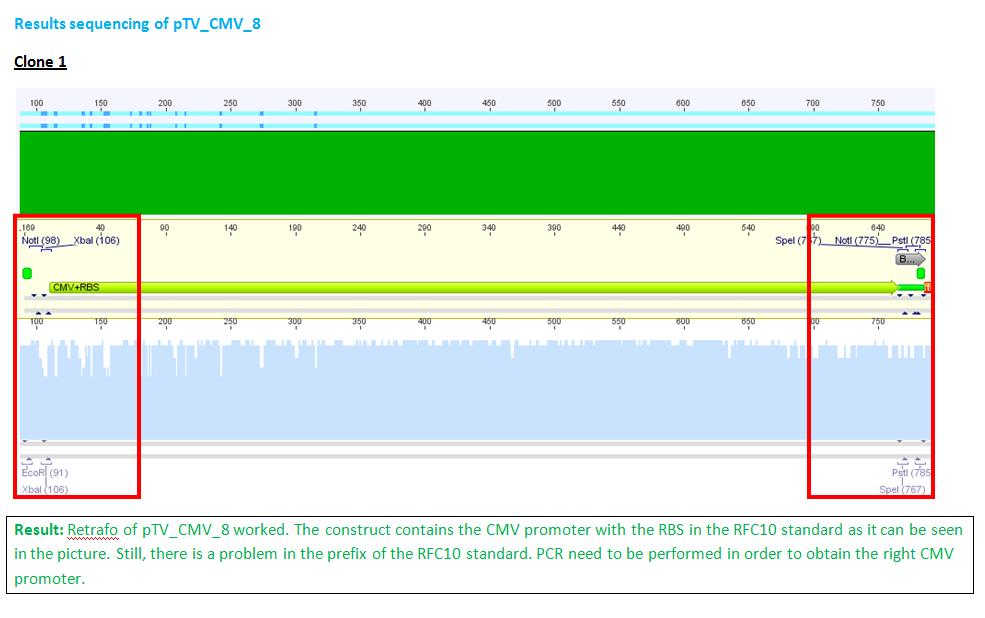
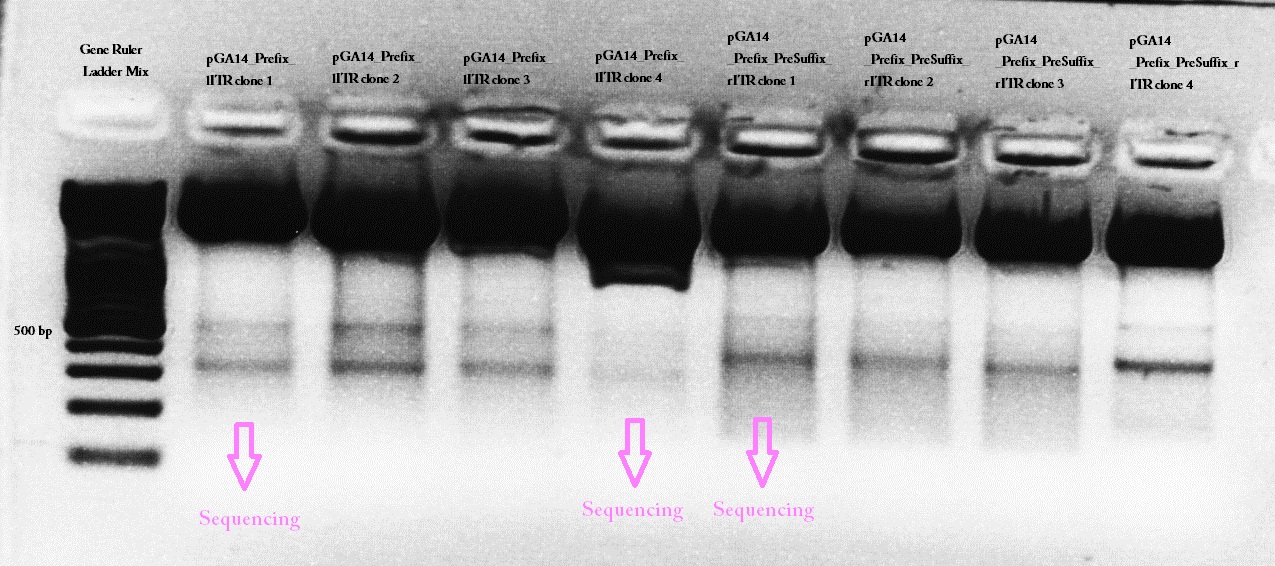
Team:Freiburg Bioware/NoteBook/Labjournal/July
From 2010.igem.org

Juli
46.Labortag 01.07.2010
qPCR
Investigators: Hanna, Chris W. (guided by Sven)
Protocol will be inserted :)
Thawing HT1080 cells
Investigators: Adrian, Bea, Patrick
- HT1080 cells were thawd following standard protocols
- HT1080 passage 3
- Incubation at 37°C
Investigation of the Kleinschmidt sequencing
Investigator: Volker
The sequence data from VM_2_30.06.-CMV-F and VM_3_30.06.-CMV-F arrived and showed as expected a homologous region with the ORFs of the corresponding VPs.
The process of VPex1, VPex2 and VPex3 sequencing is nearly finished. For further progress the oligos that prime at different locations in the Rap/Cap ORFs are required. The actual progress of the sequencing is shown in the image on the right side:
- For the sequencing of VPex1 and VPex2 one further read will be required to achieve a continuous contig.
- The sequencing of VPex3 is completed but contains two point mutations.
The investigation of the point mutations revealed that the:
- first mutation (TAC=>TAT) is silent and codes for Tyrosine. The original codon TAC has a codon usage of 58% in Homo sapiens and 25% in E. coli where as the new codon TAT has a codon usage of 42% in H. sapiens and 75% in E. coli. There for this mutation could be an optimisation of the sequence for an approach to express the VP in E.coli.
- second mutation (CCG=>CCA) is silent and codes for Proline. The original codon CCG has a codon usage of 11% in H. sapiens and 77% in E.coli weher as the new codon CCA has a codon usage of 27% in H. sapiens and 15%. This mutation would lead to an optimiced expression in human cells.
Therefor the combination of these two "optimized" codon does not make any sense in this combination.
47.Labortag 02.07.2010
Fluoroscence Pictures of Transduction 3
Investigators: Chris W, Patrick
total celldeath!
Looks like we should not transduce 3x10^5 cells with 1000 µl (or more) of our viral solution because almost all cells died in all approaches and the medium indicator was yellow.
So we can not say if resuspendig the cells with the viral soultion is a useful alteration of the standart protocol.
@Chris and Patrick: The cells which has not been resuspended died aswell?? yes (Patrick) ... so u think the volume (1000µL) of the viral stock solution is too high and cell death is not caused by the "rough" reuspension? yes (Patrick) ... Could u maybe upload some pictures even though it did not work, but to documentate it properly!! thx Bea ... No, already thrown away and we could not take good pictures because of very thick bunches of dead cells, results confirmed by Sven (Patrick)
Seeding and Splitting HEK
Investigators: Johannes, Kira, Patrick, Chris W
- 4 Petri dishes (each 10ml DMEM, 1,5*10^6 cells) and one 75 cell culture bottle (20ml DMEM, 1,5*10^6 cells). The cell density was not sufficient so we could not seed 3x10^6 in the 10 cm dishes as planed initially.
- New DMEM Medium was made (10% FCS,). We received a Sodium-Pyruvat stock solution (100x).
New ologos/primers arrived
Investigator: Patrick
ST00147058: long pITR right for, labeled with number 31
ST00147059: long pITR right rev, labeled with number 32
ST00147606: long pITR left for, labeled with number 33
ST00147061: long pITR lef rev, labeled with number 34
An additional 1:10 working dilution was made with each oligo.
@ Achim: plz write it down into our oligo excel sheet.
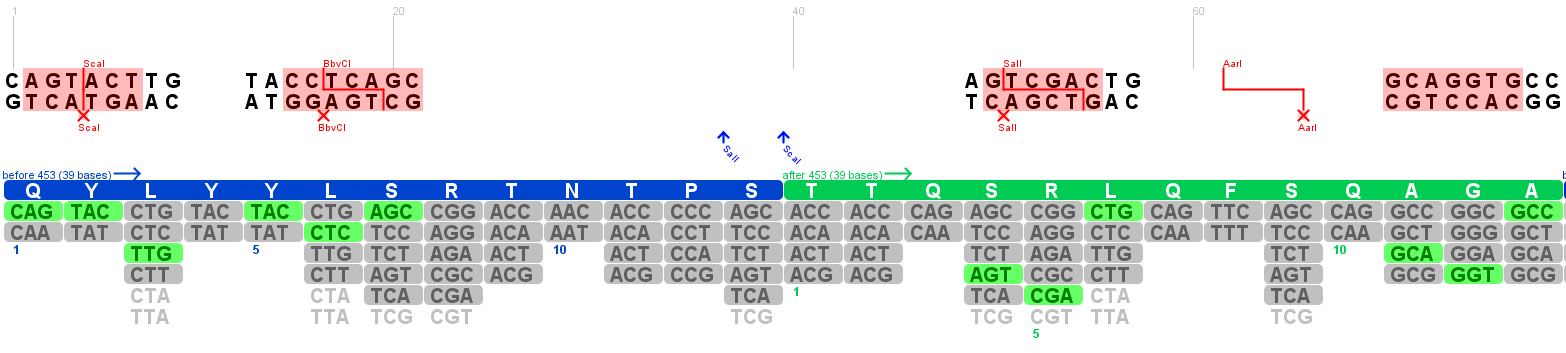
Annotation of the 453 insertion site
Investigators: Patrick & Volker
The 453 insertion site is acoording to [Boucas et al., 2009] a promising alternative to the 585/588 integration site.
The Peptides were integrated between the 453 Glycine and the 454 Threonine. This risidues were annotated in the gene maps (Genious) and visualized in the 3D structure (PyMOL).
infos from the paper
infos from the paper
infos from the paper
infos from the paper
Searching new restriction sites in the 585/588 WT loop
Investigator: Volker
The sequence of the major site for peptide integration into the capsid proteins was extracted from the viral sequence and pasted into the program DNA 2.0. In an first atempt the 4 amino acids that form the loop (5' - RGNR - 3') were used with the two flanking amino acids from the wild type sequence on each side. The goal of this theoretical investigation is to use alternative codons to insert restriction sites into the wild type loop to make later cloning easier. This was repeated in an second atempt with five flanking amino acids but did not reveal additional useful restriction sites.
Informations for the restriction enzymes were looked up in the [http://www.neb.com/nebecomm/products/category1.asp?#2 NEB Product catalogue].
The three restriction sites are:
BbvI restriction site:
Recognized sequence:
5'- GCAGC(N)8^ -3'
3'- CGTCG(N)12^ -3'
Sites found in pAAV_RC at position:
- 17 times in the ORFs of Rep & Cap => can't be used
BstEII restriction site:
Recognized sequence:
5'- G^GTNACC -3'
3'- CCANTG^G -5'
- There is only one BstEII restriction site in the vector that we want to use to insert the fragment (x-x)
- The cutting site could be removed => theoretically useful restriction site.
- Location fits nearly perfect because peptids ae usually integrated before the R585.
PstI restriction site
Recognized sequence:
5'- CTGC^AG -3'
3'- G^ACGTC -5'
- PStI is part of the iGEM standard and can't be used for this purpose.
Searching new restriction sites in the 585/588 KO loop
Investigator: Volker
PstI restriction site
Recognized sequence:
5'- CTGC^AG -3'
3'- G^ACGTC -5'
- PStI is part of the iGEM standard and can't be used for this purpose.
Summary: There is no codon combination availible to encode the promising BstEII restriction site in this R585A_R588A_KO sequence.
Searching new restriction sites in the 453 loop
Investigator: Volker
KpnI restriction site
Recognized sequence:
5'- GGTAC^C -3'
3'- C^CATGG -5'
Sites found in pAAV_RC at position:
- 1721 and 3973
- first restriction site could be replace without problem becauce it lies in the region that will be synthesized to replace the three iGEM restriction sites in the Rep ORF.
- Restriction site is potentially useful
AvaI restriction site
Recognized sequence:
5'- C^YCGRG -3'
3'- GRGCY^C -5'
Sites found in pAAV_RC at position:
- six times in the ORFs of Rep and Cap => can't be used
DraIII restriction site
Recognized sequence:
5'- TTT^AAA -3'
3'- AAA^TTT -5'
Sites found in pAAV_RC at position:
- 1181 (Rep ORF)
- 2893 (Cap ORF)
- 4463 (short after the Cap ORF)
- 4930 (f1 ORI)
AvaII restriction site
Recognized sequence:
5'- G^GWCC -3'
3'- CCWG^G -5'
Sites found in pAAV_RC at position:
- eight times in the ORFs of Rep and Cap => can't be used
Summary:
The KpnI restriction site seems to be a good possibility to make the 453 insertion site ready for easy integration of peptides.
Especially the position where the cut is performed is ideal for our purpose because peptides are usually integrated after the Glycine 453 as described for example in [Boucas et al.; 2009].
48.Labortag 03.07.2010
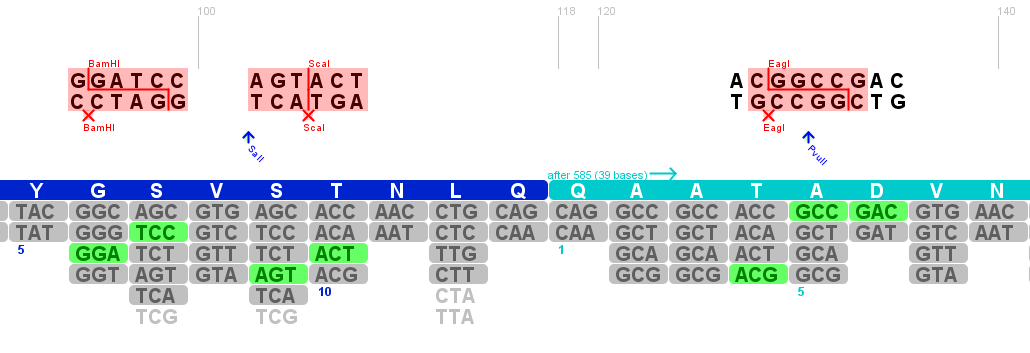
Planing the gene synthesis to replace the three restriction sites in the rep ORF
Investigator: Volker
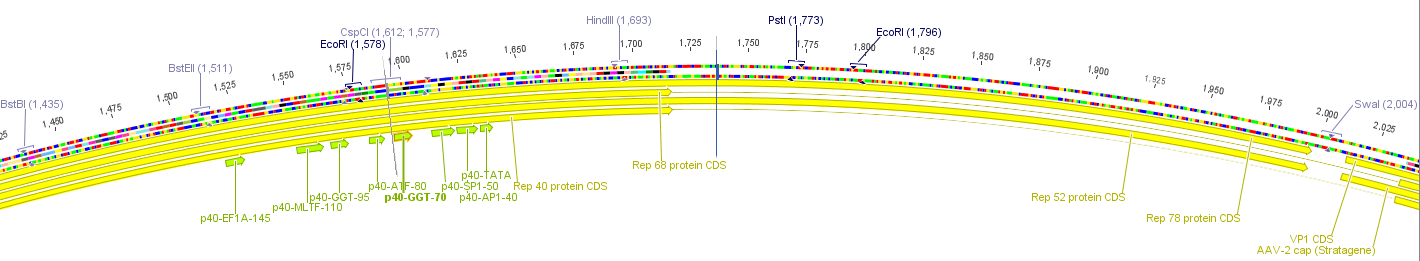
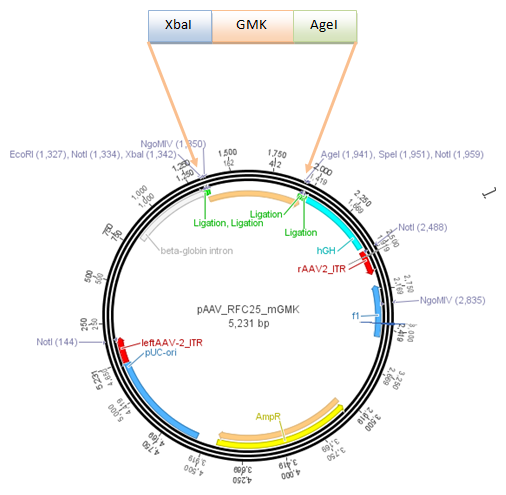
The construct pAAV_RC that we have bougth from Stratagene will be a central part of our Virus construction kit and therefor it has to be cleared from restriction sites that are used in the iGEM-standard.
There are five iGEM restriction sites in the pAAV-RC:
- PstI at position (310)
- EcoRI at position (1578)
- PstI at position (1773)
- EcoRI at position (1796)
- PstI at position (4073)
The relative location to each other and other planed modifications in this sequence make it reasonable to replace the PstI (310) and PStI (4073) with site directed mutagenesis and to order a synthesis of the region with the three other restriction sites.
The primers for the site directed mutagenesis were designed:
Designed Primer: SDM PstI (310)
- PstI at position (310)
- Recognized sequence: 5'- CTGC^AG -3'
- The codon 5'- CTG - 3' will be replaced by 5'- CTC -3'
- Both codons encode Leucine. The condon usage in H. sapiens decreases from 36% to 20%
- Recognized sequence: 5'- CTGC^AG -3'
5'- CTGACCGTGGCCGAGAAGCTCCAGCGCGACTTTCTGACGGA - 3'
Designed Primer: SDM PstI (4073)
- Recognized sequence: 5'- CTGC^AG -3'
- The codon 5'- CTG - 3' will be replaced by 5'- CTC -3'
- Both codons encode Leucine. The condon usage in H. sapiens decreases from 36% to 20%
- Recognized sequence: 5'- CTGC^AG -3'
5'- CGTGGAGATCGAGTGGGAGCTCCAGAAGGAAAACAGCAAACGCTGG -3'
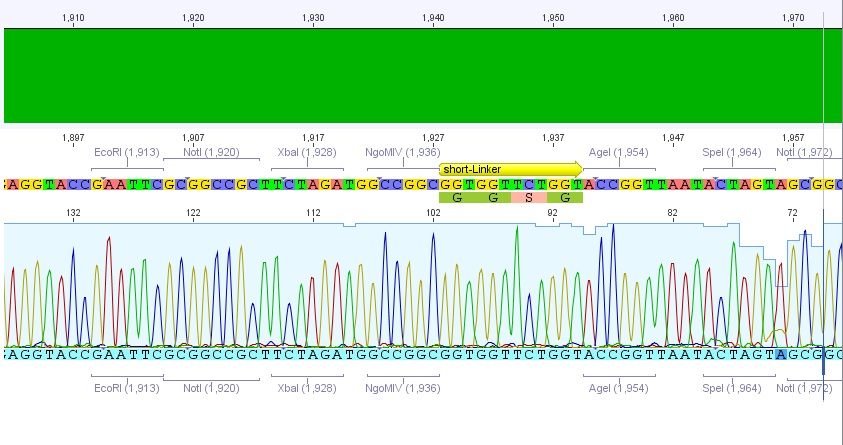
The region with the three restriction sites that are located near to each other is show in the picture above. The restriction sites indicated in light blue are single cutters and can therefor be use to clone the synthesized fragment into the original sequence of pAAV-RC.
For this purpose the restriction sites BstBI (1435) and SwaI (2004) will be used because they are availible in the lab stock and have 100% fidelity in one of the NEB buffers.
The required construct with four flanking base pairs on each side will be 583 bp long.
- EcoRI at position (1578)
- Recognized sequence: 5'- G^AATTC -3'
- The codon 5'- GAA -3' will be replaced by 5'- GAG -3'
- Both codons encode Glutamic acid. The codon usage in H.sapiens increases from 41% to 59%.
- Recognized sequence: 5'- G^AATTC -3'
- PstI at position (1773)
- Recognized sequence: 5'- CTGC^AG -3'
- The codon 5'- TGC - 3' will be replaced by 5'- TGT -3'
- Both codons encode Cysteine. The condon usage in H. sapiens decreases from 58% to 42%
- Recognized sequence: 5'- CTGC^AG -3'
- EcoRI at position (1796)
- Recognized sequence: 5'- G^AATTC -3'
- The codon 5'- AAT -3' will be replaced by 5'- AAC -3'
- Both codons encode for Asparagine. The codon usage in H. sapiens increases from 44% to 56%.
- Recognized sequence: 5'- G^AATTC -3'
This are all the restriction sites in the ORFs for the Rep and Cap proteins. In the backbone of the pAAV_RC there are more restriction sites in the iGEM standard. These restriction sites should not cause any problems when the insert in cloned into a standard iGEM vector. An other interesting question is what has been changed in the regulatory region p5. This will be important to determine the sequence that is required for the functional expression of the pAAV-RC.
Modification of the Cap genes
Investigator: Volker
The HSPG-binding-motif can be knocked out by the two mutations R585A and R588A.
The sequence 5'-CTC CAG AGA GGC AAC AGA CAA -3' codes in the pAAV-RC construct for the amino acids LQRGNRQ.
In order to achieve the recommanded mutation the sequence has to be changed to: 5'-CTC CAG GCT GGC AAC GCT CAA -3'
which codes for LQAGNAQ.
The problem is that in this R585A R588A KO version there is no possible codon usage to encode a BstEII restriction site that would make it possible to integrate peptides in an easy fashion.
An R585A R588L KO version would contain the BstEII restriction site and could be a possible alternative to the R585A R588A KO version.
Questions to adress on monday:
- Is the proposed R585A_R588L_KO mutation possible?
- Are iGEM standard vectors availible that do not contain KpnI and BstEII?
Cellculture
Investigators: Patrick, Chris W.
HT1080 cells were split. Deviations from the standart protocol: The cells were centrifuged with 2000 G instead of 200 G. Then the cells were put into the incubator for 4 hours and then examined again. It seems that most of them survived this mistake.
The 293 cells were checked for cell density. They will be >70% confluent tomorrow and therefore transfected.
49.Labortag 04.07.2010:
Transfection nr.3
Investigators: Chris W, Patrick
Transfection with P41 was performed according standart protocol.
Two transfektions where carried out with 10µg (3,33 µg each plasmid) DNA and the other two with 20µg DNA (6,66 µg each plasmid).
50.Labortag 05.07.2010:
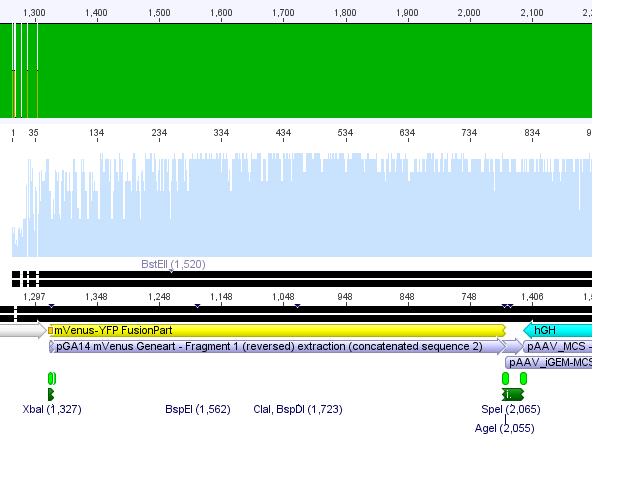
Sequencing of pAAV_iGEM-mVenus-YFP
Investigator: Hanna
Alignment of sequenced hGH region with pAAV_iGEM_mVenus-YFP delivered following picture:
There seemed to be a nucleotide insertion. But by having a closer look, we recognized that Geneious mis-interpreted the signal at this point. Further on we could validate that YFP was also successfully cloned into this clone. Unfortunately the sequencing of the gene of interest didn't work - neither with the forward, nor with the reverse primer. Their sequences were checked on the order form: Everything OK!
To do: Repeat sequencing of GOI!
Dirty and fancy method for ITR BioBrick production
Investigator: Hanna
A new strategy for ITR BioBrick production was figured out. This method does NOT require any PCR steps! :)
We want to take advantage of the fact, that the ITRs are flanked by NotI and PstI.
"Short version" of the strategy:
- Digestion of pAAV_MCS with AlwNI. Result: one large fragment (2982 bp) with right ITR; one small fragment (1674 bp) with left ITR.
- Separate fragments via gel run.
- Digest them with NotI and PstI.
- Präfix and suffix oligos were designed. They need to be hybridized and ligated to the left and right ITR with several intermediate steps (subcloning into pSB1C3 etc.). The oligos posess compatible overhangs to the PstI and NotI restriction sites of the ITR but do not generate new restriction sites after ligation!
- For detailed instruction, check up here: File:Freiburg10 ITRBioBrick dirty fancy.pdf
Beta-globin BioBrick production
Investigator: Hanna
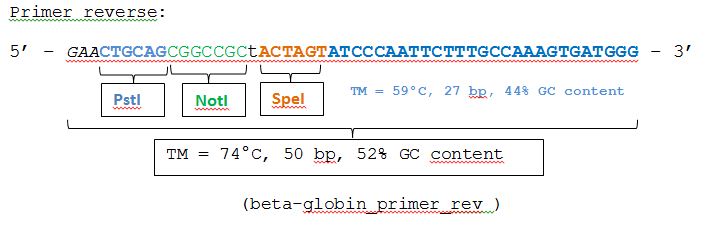
The beta-globin sequence (pAAV_MCS) contains no iGEM-restriction sites. Therefore we decided NOT to order the sequence, but to perform a PCR with RFC10 overhang primers:
hGH BioBrick production
Investigator: Hanna
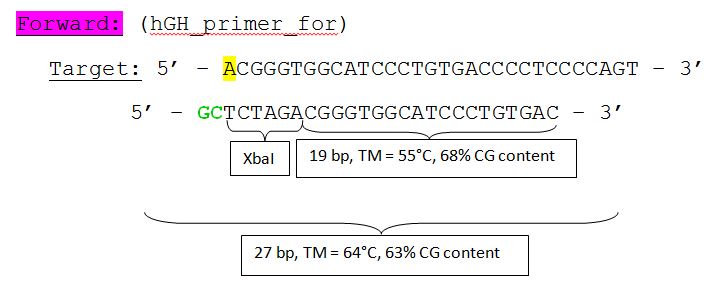
The hGH sequence (pAAV_MCS) contains no iGEM-restriction sites. Therefore we decided NOT to order the sequence, but to perform a PCR with RFC10 overhang primers:
Splitting HT and HEK cells
- Investigators: Adrian, Bea
- HEK Cells: over 100% confluent!!!
- HT-cells: over 90% confluent!!!
Preparation of HT1080 3x6er dishes for Transduction
- Investigators: Adrian, Bea
- 3x 6well dishes got prepared with 3ml DMEM and 300.000 cells (0,0375ml of our Cell-falcon)
- Transduction on 6.7.2010
Preparation of 6x10cm petri dishes HEK293
- Investigator: Adrian
- 6x 10cm petri dishes with 10ml with 3.000.000 cells (0,344ml of cell-falcon)
- Transfection on 7.7.2010
Repetition of ITR restriction site modification via PCR
- Because the previous attempt to replace the PSTI and NOTI restriction sites in the ITRs with the IGEM standard were unsucessful, new, longer primers were designed an ordered.
- a new pcr reaction with higher annealing temperatures was prepared:
| components | v(pAAV_fragment left ITR, 0% DMSO) /µl | v(pAAV_fragment left ITR, 10% DMSO) /µl | v(pAAV_fragment right ITR, 0% DMSO) /µl | v(pAAV_fragment right ITR, 10% DMSO) /µl | positive control from Gerrit: XBL PMA DBSA (106 bp) |
| 10xThermoPol buffer | 5 | 5 | 5 | 5 | 5 |
| dNTP mix | 1 | 1 | 1 | 1 | 1 |
| Primer for | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 |
| Primer rev | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 |
| DNA Template | 0,513 | 0,513 | 0,415 | 0,415 | 0,345 |
| MgSO4 | 1 | 1 | 1 | 1 | 1 |
| DMSO | 0 | 5 | 0 | 5 | 0 |
| Vent Polymerase | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 |
| H2O | 38,99 | 33,99 | 39,09 | 34,09 | 39,16 |
| Volume total | 50 | 50 | 50 | 50 | 50 |
PCR program:
| Cycles | Temperature | Time |
| 95°C | 3" | |
| 95°C | 30" | |
| 10x | 62+-2°C | 22" |
| 72°C | 12" | |
| 95°C | 30" | |
| 15x | 66+/-2°C | 22" |
| 72°C | 12" | |
| 1x | 72°C | 5' |
| Hold 4°C |
Possible reasons: *Old Vent polymerase -> New polymerase ordered
- Mg concentration could be optimized
- Template okay? Template will be loaded on gel as well next time
Search for restriction sites in the sequence LQA/RGQA/RQA
Investigator: Volker
The idea to use a R588L mutation instead of the R588A that is published in the literature was discussed with the supervisers. It was proposed to search for other amino acid exchanges that alter the loop less drastically. For this reason the mutation N587Q was examined for possible restriction sites.
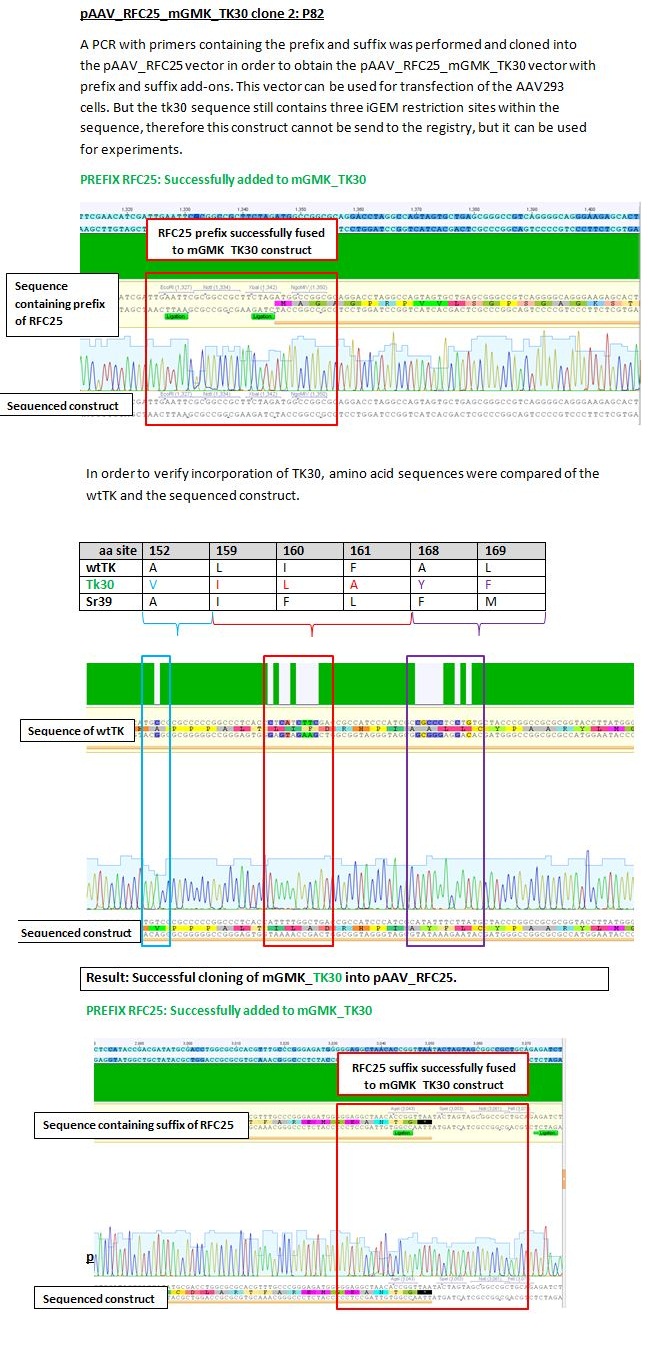
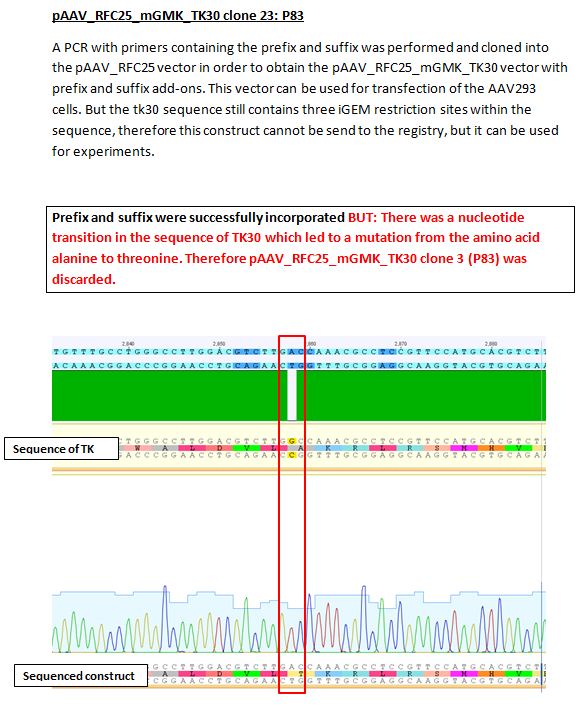
Results sequencing alignment of GMK_TK30
Investigator: Bea
Result: The obtained sequencing results indicate that the DNA we received from Amor contains the GMK_TK30 fusion enzyme, but tk30 contains some restriction sites.
- To do (06.07.2010): Order primers for BioBrick Fusionenzyme and order genes of GMK and TK (wtTK, TK30, sr39????)
- remove iGEM restriction sites
51.Labortag 6.07.2010:
Harvesting AAVs from 4 10cm dishes, Transduction of 3x6er dishes
Investigators: Adrian, Anna, Bea, Kerstin
We want to investigate which of the following approaches yields a better transduction efficiency.
- Half of the transfected cells were exposed 3 cycles of thawing and freezing and then centrifuged (15 ml falcon, 2100G). The supernatant was transfered into a new 15 ml falcon and used for transduction.
- The other half was centrifuged at 300 G for 5min . The supernatant was transfered into another 15 ml falcon and the pellet was resuspended with 5 ml of DMEM. The content of these two falcons was also exposed to 3 cycles of thawing and freezing and then used for transduction.
- We have got 2 10 cm cellculture dishes with 10µg and 2 dishes with 20µg DNA used for transfection.
Approach with standart protocol (one dish with 10 and the other with 20µg Plasmids)(2100 G and 3 cycles of freezing and thawing)
Deviations from the standart protocol:
- The cells were centrifuged at 2100 G instead of 10.000 G
- The viruses were harvested after 44 hours
- 3 cycles of freezing and thawing
Search for restriction sites before and after the insertion sites 453 and 585/588
Investigator: Volker
Transduction of 3x6er dishes
Investigators: Adrian Bea
- used plasmids: 10µg, 20µg YFP
- we have 10µg and 20µg from standart protocol => SV
- we have 10µg and 20µg from standart protocol pellet => Pellet
- we have 10µg and 20µg from standart protocol suspension => Super
- amount of 500µl
1. Plate I(A is up)
| 10µg dropped Super | 10µg dropped Super gently resuspending | 10µg dropped Super gently resuspending |
| 10µg Super gently resuspending Super | 20µg dropped Super gently resuspending 1:10 dilution | control |
2. Plate I(A is up)
| 10µg dropped Super | 10µg dropped Super gently resuspending | 10µg dropped Super gently resuspending |
| 10µg Super gently resuspending Super | 20µg dropped Super gently resuspending 1:10 dilution | control |
3. Plate I(A is up)
| 10µg dropped Pellet | 10µg dropped Pellet gently resuspending | 10µg dropped Pellet gently resuspending |
| 10µg Super gently resuspending Pellet | 20µg dropped Pellet gently resuspending 1:10 dilution | control |
Search for restriction sites around the 453 and 585/588 integration sites
Investigator: Volker
For this purpose a list of all commercially availible restriction enzymes was created that do not match in the ORFs of pAAV-RC.
Search for restriction sites in the sequence LQA/RGQA/RQA
Investigator: Volker
The idea to use a R588L mutation instead of the R588A that is published in the literature was discussed with the supervisers. It was proposed to search for other amino acid exchanges that alter the loop less drastically. For this reason the mutation N587Q was examined for possible restriction sites.
52.Labortag 07.07.2010:
Sequencing of GIO
Investigator: Hanna
Comments: Neither sequencing with forward nor with reverse primer (O29 + O30) worked. Therefore P40 (pAAV_iGEM_mVenus-YFP) was sent for sequencing once again.
- Name: Hanna P40
- V (P40) = 11 µL + 19 µL H2O
- Name of primer forward: Hanna 1 (O30)
- Name of primer reverse: Hanna 2 (O29)
- V(primer) = 3 µL + 27 µL H2O
Interview with Bild der Wissenschaft
- Adrian did a good job ;-)
- The article will be send to us next week
- For additional question, Jessica will be send an email
Transfection
Investigators: Adrian, Bea
- Transfection of AAV-293 cells with pAAV_iGEM-mVenus_YFP
New pcr run with new polymerase and different MgSO4 concentrations
- While this time the positive control was actually positive, no pcr product could be seen. The reasons are unclear. (Secondary structures, primer binding...)
Checking the results of the following sequencing reactions for the expression constructs from the DKFZ
Investigator: Volker
pKEX-VP1ex: sequence confirmed as described in [Name et al.,] with two pointmutations (ACG=>GCG) and (ATG=>CTG) to avoid the translation of VP2 and VP3.
pKEX-VP2ex: sequence confirmed as described in [Name et al.,] with one mutation (ATG=>CTG) to avoid the translation of VP3 and a second mutation (AAGACG=>CATATG) to switch to the main start codon ATG and to introduce a restriction site (which one??) before the start codon.
pKEX-VP3ex: sequence confirmed as described in [Name et al.,] with two silent pointmutations and a second mutation to introduce a restriction site before the start codon.
pKEX-Rep40ex: Sequence as expected until now, further sequencing reaction required with the primer: GATC_std_pTeSp-1
pKEX-Rep52ex: Point mutation directly at the beginning of the exon sequence, perhaps functionality in this context? At the end of the sequence some umcertainities. For the beginning further sequencing reaction required with the primer: GATC_std_pTeSp-1
pKEX-Rep68ex: One point mutation in the second condon that changes the coded amino acids from P to A, a second mutation (ATG=>GGG) at the beginning of the Rep40/Rep52 ORFs to avoid the translation of these sequences and a silent mutation. One further sequencing reaction is required with the primer: GATC_std_pcDNA1.1-RP.
pKEX-Rep78ex: One point mutation in the second condon that changes the coded amino acids from P to A, a second mutation (ATG=>GGG) at the beginning of the Rep40/Rep52 ORFs to avoid the translation of these sequences, a silent mutation and the same point mutation at the beginning of the exon as seen before in pKEX-Rep52ex.
53.Labortag 08.07.2010:
Final planing of the Cap modifications
Investigator: Volker
BbvCI
50|100|10|100 37°
File:Freiburg10 BbvC-I-cutsite 1 v1 000007.gif
Recognition sites in Rep/Cap ORFs:
Recognition sites in pAAV_RC:
Recognition site in pSB1C3:
KpnI
100|75|0|50 37°C + BSA
File:Freiburg10 Kpn-I-cutsite 1 v1 000021.gif.gif
Recognition sites in Rep/Cap ORFs:
Recognition sites in pAAV_RC:
Recognition site in pSB1C3:
SalI
10|100|100|100 37°C
File:Freiburg10 Sal-I-cutsite 1 v1 000016.gif.gif
Recognition sites in Rep/Cap ORFs:
Recognition sites in pAAV_RC:
Recognition site in pSB1C3:
AarI
|||
File:Freiburg10 .gif
Recognition sites in Rep/Cap ORFs:
Recognition sites in pAAV_RC:
Recognition site in pSB1C3:
ScaI
100|100|10|100 37°C
File:Freiburg10 Sca-I-cutsite 1 v1 00001.gif.gif
Recognition sites in Rep/Cap ORFs:
Recognition sites in pAAV_RC:
Recognition site in pSB1C3: 2
BamHI
75|100|100|100 37°C + BSA
File:Freiburg10 BamH-I-cutsite 1 v1 000025.gif.gif
Recognition sites in Rep/Cap ORFs:
Recognition sites in pAAV_RC:
Recognition site in pSB1C3:
PvuII
100|100|100|100 37°C
File:Freiburg10 Pvu-II-cutsite 1 v1 000013.gif.gif
Recognition sites in Rep/Cap ORFs:
Recognition sites in pAAV_RC:
Recognition site in pSB1C3: 1
EagI
10|25|100|10 37°C
File:Freiburg10 Eag-I-cutsite 1 v1 000021.gif.gif
Recognition sites in Rep/Cap ORFs:
Recognition sites in pAAV_RC:
Recognition site in pSB1C3: 2
54.Labortag 09.07.2010:
Investigators: Anissa, Kerstin, Anna
- New DMEM-Medium was prepared (10% FCS). Investigators: Bea, Patrick
- HT and 293 cells were split into 4 flasks, 2 flasks each. Investigator: Kira
- AAV stocks were prepared, put into the -80°C freezer and labeled with the date, name, AAV, the amount of DNA used for transfection and drop/resuspend to make clear whether the DNA was just pippeted dropwise to the 293 cells or gently mixed with the cells. The numbers on the 15 ml falcons refer to DNA amount and DNA administration, too. Investigator: Patrick
PCR of hgH
- PCR program: Unnamed
| components | volume of vector /µl |
| 5x Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| hgH_primer_for | 2,5 |
| hgH_primer_rev | 2,5 |
| DNA template | 0,38 |
| DMSO | - |
| Phusion polymerase | 0,5 |
| H2O | 33,1 |
| Total volume (e.g. 50 µl) | 50 |
Agarose-Gel:
0,625 g Agarose, 50 ml TAE (1,25%), 3 µl GELRED (3-6µl), at 110 Volt, running time: 60 minutes
| Sample | Sample/µl] | Loading dye (5x/6x)/µl | Expected size (Geneious) |
|---|---|---|---|
| p47: hgH | 50 µl | 10 µl | 488 bp |
- Marker: GeneRuler ladder mix
| Marker (8 µl) | Sample 50 /µl | |
|---|---|---|
| Lane | 1 | 4 |
Gelextraction
Gel measurement:
| Sample | weight |
| p47: hgH | 0,13 g |
- DNA concentration (hgH): 19,83 ng/µl
Comments: PCR was succesful:-)
to do: Ligation of PCR product into standard plasmid which we´ve received wihthin the DNA distrivution kit of iGEM-HQ .
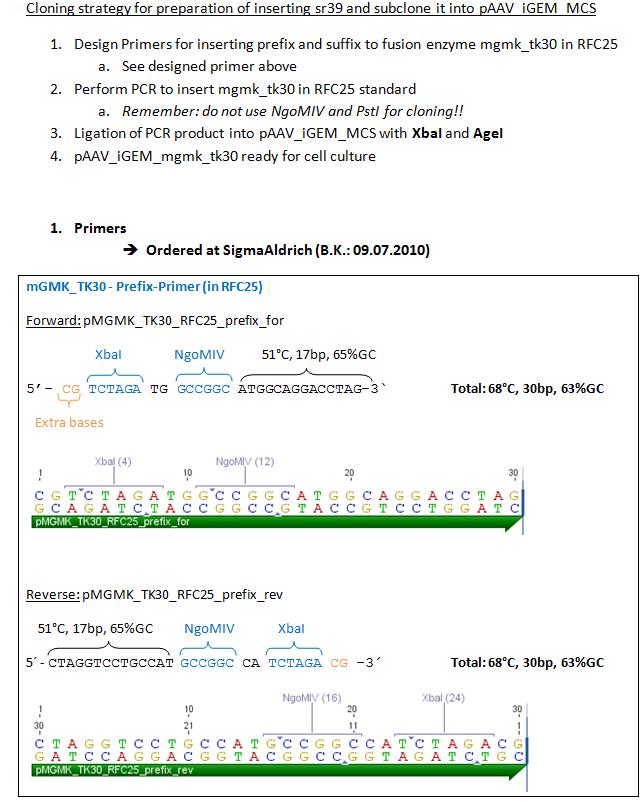
Order oligos for GMK_TK30
Investigator: Bea
Oligos have been ordered at sigmaaldrich in order to insert prefix and suffix to fusionenzyme mGMK_TK30 in RFC25.
to do: Order gene tk30 and part of gene sr39
Quickchange for the PstI restriction sites in pAAV-RC
Investigators: Kira and Volker
Quick change, Gel and test digestion with PstI. (more precise informations will be added)
@Kira and Volker: did the Quickchange actually work? Could you please add the information onto the wiki!! Thx!
PCR program: PstI
| components | volume in µl |
| 10x Phusion HF buffer | 2.5 µl |
| 10 mM dNTP mix | 0.5 µl |
| __310 ____primer_for (1:10 dilution) | 0,39 µl |
| __310____primer_rev (1:10 dilution) | 0,39 µl |
| DNA template (1:40 dilution) | 0,5 µl |
| DMSO | 0 µl |
| Phusion polymerase | 0.5 µl |
| H2O | 20,22 µl |
| Total volume (e.g. 50 µl) | 25 µl |
| components | volume in µl |
| 10x Phusion HF buffer | 2.5 µl |
| 10 mM dNTP mix | 0.5 µl |
| __4073____primer_for (1:10 dilution) | 0.33 µl |
| ___4073___primer_rev (1:10 dilution) | 0.33 µl |
| DNA template (1:40 dilution) | 0.5 µl |
| DMSO | 0 µl |
| Phusion polymerase | 0.5 µl |
| H2O | 20.32 µl |
| Total volume (e.g. 50 µl) | 25 µl |
| cycles | temperature | time |
| 1 | 95 C | 2 min |
| 20 | 95 C | 30 sec |
| 66 C | 1 min | |
| 68 C | 5 min |
- The PCR-products were digested with DpnI (1µl instead of 0.5 µl)
- aragose gel revealed for both samples very distinct bands
TEST DIGESTION
| components | volume /µl |
| DNA | 10 |
| BSA (10x) | 2 |
| Buffer 3 (10x) | 2 |
| Enzyme (no.Lab: 48) | 1 |
| H2O | 5 |
| Total volume | 20 |
- agarose gel was performed with digested samples and revealed 3 distinct bands, while only 1-2 bands have been expected. Because of the density of the bands it might be that the digestion with DpnI did not work properly, thus additional 1 µl of DpnI was added to each sample. Incubation @ 37 C lasted 1 h.
55.Labortag 11.07.2010
Splitting of HT1080 cells
Investigator: Patrick
HT 1080 and HEK 293 cells were split.
@ Kira: fetch new trypsin if it is not sufficient for the next splitting !
56.Labortag 12.07.2010:
ITR BioBrick production
Practical Cloning:
- experiment date: 12.07.2010 ; time: 11.00
- name of investigator: Anna, Kerstin, Anissa
- plasmid: pSB1c3_CFP (p48)
- Vector: name: pSB1c3 number:- production date: - origin: iGEM
- Insert: name: pMA_CFP number:- production date: - origin: Sven
- new vector name: pSB1c3_CFP (p48)
- buffer used: 3 ; Restriction-enzymes used: Enzyme 1 (no. Lab:28) EcoRI ; Enzyme 2 (no.Lab:48) PstI ;Enzyme 3 (no.Lab:77) DpNI
- DNA concentration (vector): 25 µg/µl ; DNA concentration (insert): 436,8 µg/µl
Comments:Third enzyme was only used to digest template from vector pcr
Digestion
| components | volume of vector /µl | volume of insert /µl |
| DNA | 4 | 5 |
| BSA (100x) | 0,2 | 2 |
| Buffer ____ (10x) | 1,5 | 2 |
| Enzyme 1 (no.Lab:23) | 0,5 | 1 |
| Enzyme 2 (no.Lab:48) | 0,5 | 1 |
| Enzyme 3 (no.Lab:77) | 0,5 | 1 |
| H2O | 7,8 | 9 |
| Total volume (e.g. 15,20,25,30 µl) | 15 | 20 |
- Incubation: 1h
Agarose-Gel:
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED (3-6µl), at 110 Volt, running time:45 minutes
| Sample | Sample/µl] | Loading dye (5x/6x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) | Expected size 3 (Geneious) | Expected size 4 (Geneious) |
|---|---|---|---|---|---|---|
| 2 | 20 µl | 3,3 µl | 732 bp (Insert) | 2074 bp (Vector) | ____ bp | ____ bp |
- Marker: GeneRuler ladder mix
| Marker | Sample 23,3 µl | |
|---|---|---|
| Lane | 1 | 3 |
Gelextraction
Gel measurement:
| Sample | weight |
| 2 (Insert) | 0,14g |
Ligation
| Insert | Vector | |
| Volume/µl | 6,77 µl | 2,23 µl |
TRAFO-EVALUATION
- Unfortunately the agar plate with p48, pSB1c3_CFP [XL1B, Cm, A.B.] does not contain any colonies. Thus, the plate was incubated @ 37 C for another 5 hours. Still no detectable colonies. This was double checked by Sven.
Seeding HT1080 + Passage
- Investigators: Adrian, Patrick
- 3* 6well dishes
| 3*10^5 cells | 3*10^5 cells | 3*10^5 cells |
| 3*10^5 cells | 3*10^5 cells | 3*10^5 cells |
- The amount of cells was doubled in first place and diluted in following steps (for further information ask patrick and adrian)
PCR of beta-globin:
- Investigator: Achim
- PCR program: BETA...(?)
| components | volume of vector /µl |
| 5x Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| beta-globin_primer_for | 2,5 |
| beta-globin_primer_rev | 2,5 |
| DNA template | 8 ng |
| DMSO | - |
| Phusion polymerase | 0,5 |
| H2O | 33,2 |
| Total volume (e.g. 50 µl) | 50 |
Agarose-Gel:
0,625 g Agarose, 50 ml TAE (1,25%), 3 µl GELRED (3-6µl), at 115 Volt, running time: 30 minutes
| Sample | Sample/µl] | Loading dye (5x/6x)/µl | Expected size (Geneious) |
|---|---|---|---|
| p49: beta-globin | 50 µl | 10 µl | 509 bp |
- Marker: GeneRuler ladder mix
| Marker (8 µl) | Sample 50 /µl | |
|---|---|---|
| Lane | 1 | 4 |
Gelextraction
Gel measurement:
| Sample | weight |
| p49: hgH | 0,21 g |
- DNA concentration (hgH): 34,87 ng/µl
Comments: PCR was succesful:-)
to do: Ligation of PCR product into standard plasmid which we´ve received wihthin the DNA distrivution kit of iGEM-HQ .
Midi-Prep preparation pAAV_RC
Investigator: Bea
- 200mL LB-Medium and 200µl Amp
- inoculated with glycerol stock containing pAAV_RC
- 20:00 p.m.: put intp 37°C room on shaker
First steps in producing ITR BioBricks via Fancy Method
Investigators: Hanna, Chris W.
Digestion of pGA14_Cerulean-CFP
1. Digestion with EcoRI and NotI:
| components | volume of vector /µl |
| DNA | 3.5 |
| BSA (10x) | 2 |
| Buffer 3 (10x) | 2 |
| Enzyme EcoRI (no.Lab:23) | 1 |
| Enzyme NotI (no.Lab:46) | 1 |
| H2O | 10.5 |
| Total volume | 20 |
Incubation: 1.5 hours, 37°C
Gel run: 20 µL sample + 4 µL loading dye (6x); 8µL marker
Expected fragment size: ~ 2900 bp
Gel extraction:
Sample weight: 0.15 g
Gel extraction was performed following the standard protocol.
DNA concentration: 26.8 ng/µL
1. Digestion with XbaI and PstI:
| components | volume of vector /µl |
| DNA | 3.5 |
| BSA (10x) | 2 |
| Buffer 3 (10x) | 2 |
| Enzyme XbaI (no.Lab:63) | 1.25 |
| Enzyme PstI (no.Lab:48) | 0.75 |
| H2O | 10.5 |
| Total volume | 20 |
Incubation: 1.5 hours, 37°C
Gel run: 20 µL sample + 4 µL loading dye (6x); 8µL marker
Expected fragment size: ~ 2900 bp
Gel extraction:
Sample weight: 0.17 g
Gel extraction was performed following the standard protocol.
DNA concentration: 22.9 ng/µL
Digestion of pAAV_MCS (P9) with AlwNI
| components | volume of vector /µl |
| DNA | 11.6 |
| BSA (10x) | - |
| Buffer 4 (10x) | 2.5 |
| Enzyme AlwNI (no.Lab:120) | 2.5 |
| H2O | 8.4 |
| Total volume | 25 |
Incubation: 1 hours + 20 minutes, 37°C
Gel run: 20 µL sample + 4 µL loading dye (6x); 8µL marker
Expected fragment sizes: 2982 bp = right ITR and 1674 bp = left ITR
Gel extraction:
- Sample: right ITR = 0.15 g
- Sample: left ITR = 0.12 g
Gel extraction was performed following the standard protocol.
DNA concentration:
- right ITR: 44.5 ng/µL
- left ITR: 32.8 ng/µL
Digestion of ITR fragments with NotI and PstI
Digestion of both fragments was performed as follows:
| components | volume of vector /µl |
| DNA | 19 µL |
| BSA (10x) | 1.5 |
| Buffer 3 (10x) | 2.5 |
| Enzyme NotI (no.Lab:46) | 1 |
| Enzyme PstI (no.Lab:48) | 1 |
| H2O | - |
| Total volume | 25 |
Incubation: 1 hours + 15 minutes, 37°C
Gel run:
- due to the small fragment sizes, we prepared a 2% agarose gel: 1.3 g agarose + 65 ml TAE buffer
- 25 µL of each sample + 5 µL loading dye (6x); 8µL marker
- Expected fragment sizes: ~ 150 bp (both)
- Big problem: Small fragments and little amounts -> 150 bp fragments were nearly not visible. In addition to that, the right ITR sample delivered 2 fragments in the referring range. Therefore two gel extractions ("right ITR oben" and "right ITR unten") were performed.
Gel extraction:
- Sample: right ITR oben = 0.11 g
- Sample: right ITR unten = 0.12 g
- Sample: left ITR = 0.2 g
Gel extraction was performed with special columns provided from Sven. In order to elute the samples 12 µL EB were used, columns were placed for 2 minutes into 50°C water bath and then centrifuged at 13200 rpm for 1 minute.
DNA concentration:
- right ITR oben: 23.9 ng/µL
- right ITR unten: 13.8 ng/µL
- left ITR: 4.46 ng/µL
Hybridization of ITR-Präfix-Oligos
left ITR:
- 10 µL Oligo 1 (O43 = Präfix_for)
- 10 µL Oligo 2 (O44 = Präfix_rev)
- 4 µL 100 mM TrisHCl pH8
- 8 µL 5 mM MgCl2
- 8 µL H2O
Total: 40 mL
right ITR:
- 10 µL Oligo 1 (O47 = Präfix_for)
- 10 µL Oligo 2 (O48 = Präfix_rev)
- 4 µL 100 mM TrisHCl pH8
- 8 µL 5 mM MgCl2
- 8 µL H2O
Total: 40 mL
Because some of the oligos produce strong secondary structures, we used the ORIGAMI1 program:
1) initial denaturation: 99°C, 7 minutes
2) 99°C, 1 minute
3) 73 x repetition of 2) -> -1°C, R = 0.3 °C/sec
4) hold 4°C
Ligation of pGA14 + ITR + Präfix
1. right ITR oben:
- Präfix: 1 µL
- ITR: 7 µL
- vector (pGA14): 9 µL
- buffer: 2 µL
- T4 ligase: 1 µL
Total volume: 20 µL
2. right ITR unten:
- Präfix: 1 µL
- ITR: 7 µL
- vector (pGA14): 9 µL
- buffer: 2 µL
- T4 ligase: 1 µL
Total volume: 20 µL
3. left ITR:
- Präfix: 1 µL
- ITR: 7 µL
- vector (pGA14): 9 µL
- buffer: 2 µL
- T4 ligase: 1 µL
Total volume: 20 µL
Samples will be incubating over night at 16°C.
Ordering genes for the replacement of Rep and Cap parts
Investigator: Volker guided by Sven & Tobi
The replacement for the Rep ORF from base 1510 to base 2008 and for the Cap ORF from base 3064 to base 3985 was ordered from Mr. Gene.
File:Freiburg10 Rep Cap Replacement.pdf
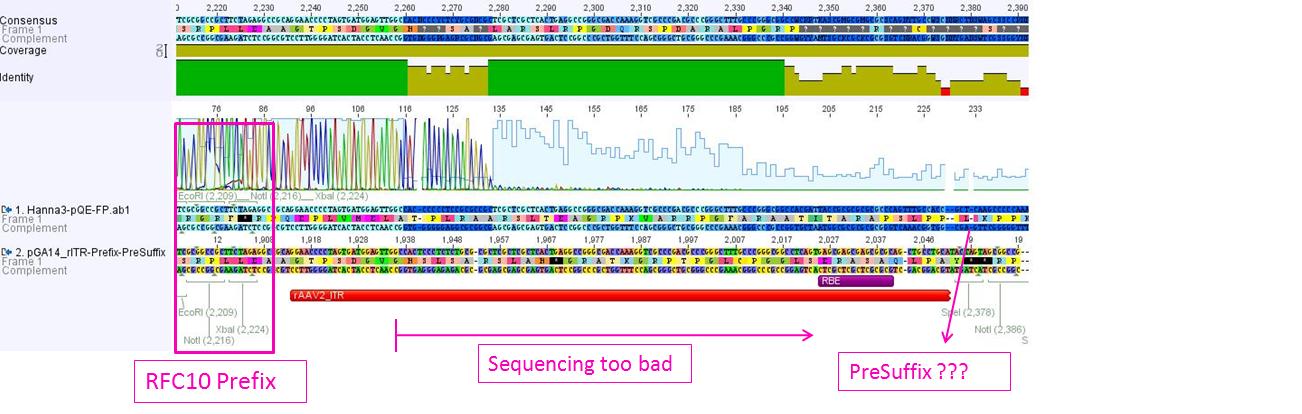
Sequencing of GOI
Investigator: Hanna
This time the sequencing delivered good results:
Conclusion: The designed primers can always be used in order to sequence any gene of interest!!!
57.Labortag 13.07.2010:
Endotoxin-free Midi Prep of pAAV_RC
Investigators: Chris, Stefan, Patrick
Deviations from the Standard Protocol:
As the lysate was applied to the column filter, about 1/5 of the lysate was spilled.
Result of the Midi-prep: about 18 ng/µl => waste
A new Midi-Pred will be prepared for tomorrow: 200 ml LB, 0,2 ml Amp, E.coli XL-1-blue (with pAAV_RC plasmid).
HEK-293 cells
The cells were split according to the standard protocol. Investigator: Patrick
Oligos pMGMK_TK30 arrived
New Oligos arrived for inserting RFC25 to thymidinkinase_gmk construct!(number O60 to O65). They were resuspended in water and a 1:10 dilution was done. They were put in Primer Box 2.
to do: perform PCR tomorrow and clone PCR product into pAAV_RFC25
Transduction
Investigators: Adrian, Patrick
- 3 Plates got transduced
1. Plate I(A is up) completely 10 µg
| 300µl dropped | 300µl resuspended | 450µl resuspended |
| 600µl dropped | 600µl resuspended | control |
2. Plate II(A is up)completely 18 µg
| 300µl dropped | 300µl resuspended | 450µl resuspended |
| 600µl dropped | 600µl resuspended | control |
3. Plate III(A is up) completely 24 µg
| 300µl dropped | 300µl resuspended | 450µl resuspended |
| 600µl dropped | 600µl resuspended | control |
Results of Trafo of cloning CFP into pSB1C3
Investigator: Bea
Results: no clones on trafo plate even after longer incubation at 37 °C
Solution: perform digestion of linearized plasmid pSB1C3 and pMA_CFP again!!!
Repetition of cloning of CFP into pSB1C3
- experiment date: 13.07.2010 ; time: 16:00
- name of investigator: Bea
- Vector: name: pSB1c3 number: do not have any number production date: - origin: iGEM HQ sent per DNA distribution kit
- Insert: name: pMA_CFP number:- production date: - origin: iGEM 2009
- new vector name: pSB1c3_CFP (P48)
- buffer used: 3 ; Restriction-enzymes used: Enzyme 1 (no. Lab: 23) EcoRI ; Enzyme 2 (no.Lab: 48) PstI ;Enzyme 3 (no.Lab: 77) DpnI
- DNA concentration (vector: pSB1C3): 25 ng/µl ; DNA concentration (insert pMA_CFP): 436,8 µg/µl
Comments:The standard plasmid vector (pSB1C3) received from the iGEM HQ is linearized (PCR product) and need to be cut with EcoRI, PstI and optionally with DpnI to digest the DNA template.
Digestion of pSB1C3 and pMA_CFP
| components | volume of pSB1C3 /µl | volume of pMA_CFP /µl |
| DNA | 4 | 4,6 |
| BSA (100x) | 1,5 | 2 |
| Buffer 3 (10x) | 1,5 | 2 |
| Enzyme EcoRI (no.Lab:23) | 0,5 | 1 |
| Enzyme PstI (no.Lab:48) | 0,5 | 1 |
| Enzyme DpnI (no.Lab:77) | 0,5 | - |
| H2O | 6,5 | 9 |
| Total volume (e.g. 15,20,25,30 µl) | 15 | 20 |
| Incubation | 30 min at 37°C in PCR cycler | 45min at 37°C in PCR cycler |
| commment | after digestion: 80°C 20 min heat kill | load on agarose gel |
- Load pMA_CFP digestion reaction onto 1% agarose gel
1% Agarose-Gel
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED (3-6µl), at 110 Volt, running time:45 minutes
| Sample | Sample/µl] | Loading dye (/6x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| pMA_CFP | 20 µl | 4 µl | 732 bp (Insert) | 2974 bp (Vector) |
Loading plan for agarose gel:
Marker used: GeneRuler ladder mix (Fermentas)
| Marker | Sample 24 µl | |
|---|---|---|
| Lane | 1 | 3 |
Results: Expected bands too high, but lower band (= CFP) was cut out and gel extraction have been performed folowing standard protocol.
Gel extraction
has been performed and Nanodrop measurement reveals following concentraion of the insert CFP:- c(CFP)= 6,75ng/µL
- Ligation ratio of vector/insert was calculated with LabTools
- v(pSB1C3)= 4,4 µL
- v(CFP)= 4,6 µL
- vQuickligase Buffer (2x)= 10µL
- vQuickLigase = 1µL
Transformation
- used bacterial strain: XL-1B
- 2µL of DNA (pSB1C3_CFP)
- Trafo plate in 37°C room for incubating at 23:00
TRANSFORMATION of Quickchange PstI
Investigator: Kira
- Transfection was performed according to the standard protocol with XL1B cells.
58.Labortag 14.07.2010
Trafo evaluation of Quickchange PstI
Investigator: Kira
Both plates contained lots of colonies, while 8-10 colonies are considered for inoculation to perform a mini prep and an additional test digestion.
However, the experiment has to be repeated.
Quickchange for the PstI restriction sites in pAAV-RC (modified)
Investigators: Kira
PCR program: PstI
| components | volume in µl |
| 10x Phusion HF buffer | 2.5 µl |
| 10 mM dNTP mix | 0.5 µl |
| __310 ____primer_for (1:10 dilution) | 0,39 µl |
| __310____primer_rev (1:10 dilution) | 0,39 µl |
| DNA template (1:40 dilution) | 0,5 µl |
| DMSO | 0.5 µl |
| Phusion polymerase | 0.5 µl |
| H2O | 19.72 µl |
| Total volume (e.g. 50 µl) | 25 µl |
| components | volume in µl |
| 10x Phusion HF buffer | 2.5 µl |
| 10 mM dNTP mix | 0.5 µl |
| __4073____primer_for (1:10 dilution) | 0.33 µl |
| ___4073___primer_rev (1:10 dilution) | 0.35 µl |
| DNA template (1:40 dilution) | 0.5 µl |
| DMSO | 0.5 µl |
| Phusion polymerase | 0.5 µl |
| H2O | 19.82 µl |
| Total volume (e.g. 50 µl) | 25 µl |
| cycles | temperature | time |
| 1 | 95 C | 2 min |
| 20 | 95 C | 30 sec |
| 68 C | 7 min |
- PCR products were digested with 0.5 µl and incubated @ 37 C for 1 h
TRANSFORMATION
- The transformation of XL1B was performed according to the standard protocol but with DYT instead of LB.
Results of trafo cloning pSB1C3_CFP
Investigator: Bea
Comments: experiment have been repeated because no clones were grown after the last trafo!
- There have been clones grown on the trafo plate. Four clones were picked and inoculated on LB_Medium containing Cm (Chloramphenicol).
- 18:00: 37°C room on shaker. Incubating over night!
Seed AAV-293
Investigators: Adrian ,Bea
- 2x75cm2 culture flasks
- after counting via neubauer chamber (2,2*20*10000*(x/4))= should be around 3,xxx so it would be possible to prepare 4x10cm petri dishes with 3.000.000 cells per plate and passage 1.500.000 cells to a 75cm.
DAAD Timetable
Investigators: Adrian, Kerstin Prepared timetable for DAAD
Summary
Investigators: all
have not been done yet! to do for tomorrow! deadline of submitting project descrpition is 16th of july!! (Bea)
Ordering ITR+TK/GMK
Investigators: Bea, Sven
- It has been ordered the TK30 with NgoMIV add-on prefix and ageI and SpeI add-on suffix combined with the wt_left_ITR in the RFC10 standard.
Endotoxoin free Midi prep mAAV_RC
Investigator: Chris W.
Midi prep done with the Qiagen-Kit
OD of the Bacteria: 3.07
concentration: 429,8 and 378,5 ng/µl
Labeled as P50 and stored in the -20° Freezer
MGMK_TK30:PCR with iGEM restriction-sites and cloning into pAAV_RFC25
Investigators: Achim,Bea,Anissa
PCR:
- PCR program:mGMK_TK30
| components | volume in µl |
| 5x Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| pMGMK_TK30_RFC25_prefix_for (060) (1:10 dilution) | 2,5 (0,5 µM) |
| pMGMK_TK30_RFC25_suffix_rev (063) (1:10 dilution) | 2,5 (0,5 µM) |
| DNA template | ~8 ng |
| DMSO | - |
| Phusion polymerase | 0,5 |
| H2O | fill up to 50 µl |
| Total volume (e.g. 50 µl) | 50 |
Comments:Oligos with add-on tails (prefix and suffix), but TK30 still contains three iGEM restriction sites (TK30 without restriction sites was ordered today)
Practical Cloning:
- Vector: name:pAAV_RFC25 number:p42
- Insert: name: MGMK_TK30 (after PCR with iGEM standard) number:-
- new vector name: pAAV_RFC25_MGMK_TK30
- buffer used:4 ; Restriction-enzymes used: Enzyme 1 (no. Lab:149) AgeI ; Enzyme 2 (no.Lab:63) XbaI
- DNA concentration (vector):321,5 ng/µl ; DNA concentration (insert):
Comments:_____
Digestion
| components | volume of vector /µl | volume of insert (=PCR product) /µl |
| DNA | 3,1 (1 µg) | 30 (0,691 µg) |
| BSA (100x) | 2 | 5 µl |
| Buffer 4 (10x) | 2 | 5µl |
| AgeI (no.Lab:149) | 1 | 2 µl |
| XbaI(no.Lab:63) | 0,75 | 1 µl |
| H2O | 11,15 | 7 µl |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 50 µl |
- Incubation: Vector: 1h; PCR product: 2h
Agarose-Gel:
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED (3-6µl), at 110 (PCR); 115 (vector) Volt, running time:____ minutes
| Sample | Sample/µl] | Loading dye (5x/6x)/µl | Expected size (Geneious) |
|---|---|---|---|
| PCR product | 50 µl | 10 µl | ~1692 bp |
| vector | 20 µl | 4 µl | 4646 bp |
- Marker: GeneRuler ladder mix
| Marker | Sample 60 /µl | |
|---|---|---|
| Lane | 8 µl | PCR |
- Marker: GeneRuler ladder mix
| Marker | Sample 24 /µl | |
|---|---|---|
| Lane | 8 µl | vector |
Comments: two gels were made, one for the vector, one for the pcr product
- Gel extraction was carried out according to standard protocol
- Nanodrop:
- pAAV_rfc 25 Digestion: 15,07 ng/µl
- mgmk tk30 pcr product: 23,04 ng/µl
- Puricifation using Quiagen QIAquick PCR Purification Kit Protocol
- c(mgmk_tk30) = 8,1ng/µL
- Ligation of mgmk tk30 and pAAV_rfc 25
- v(vector) = 2,97 µL
- v(insert) = 6,03 µL
- Trafo
- put in 37°C room at 22:00
Ordering of Oligos
Investigator:Volker
The following four Oligos were ordered:
- SDM BamHI (729) for
- SDM BamHI (729) rev
- SDM SalI (1110) for
- SDM SalI (1110) rev
59.Labortag 15.07.2010
Picking clones for mini-prep
Investigators: Adrian, Chris, Stefan
4 clones (1-4) of pAAV_RFC25_mgmk_tk30 where picked according to protocol for mini-prep (tomorrow).
Antibiotic: Amp
Time: 18:30
Investigators: Stefan, Patrick
New millipore H2O, LB + Agar and DMEM medium were prepared
Splitting of HT1080 cells
Investigators: Chris, Patrick
HT 1080 cells were split according to the standard protocol
PstI Quickchange Trafo from 14.07
Investigator: Kira
Trafo evaluation: Unfortunately no colonies are present.
MINI PREP (PstI Quickchange Trafo from 13.07)
Investigator: Kira
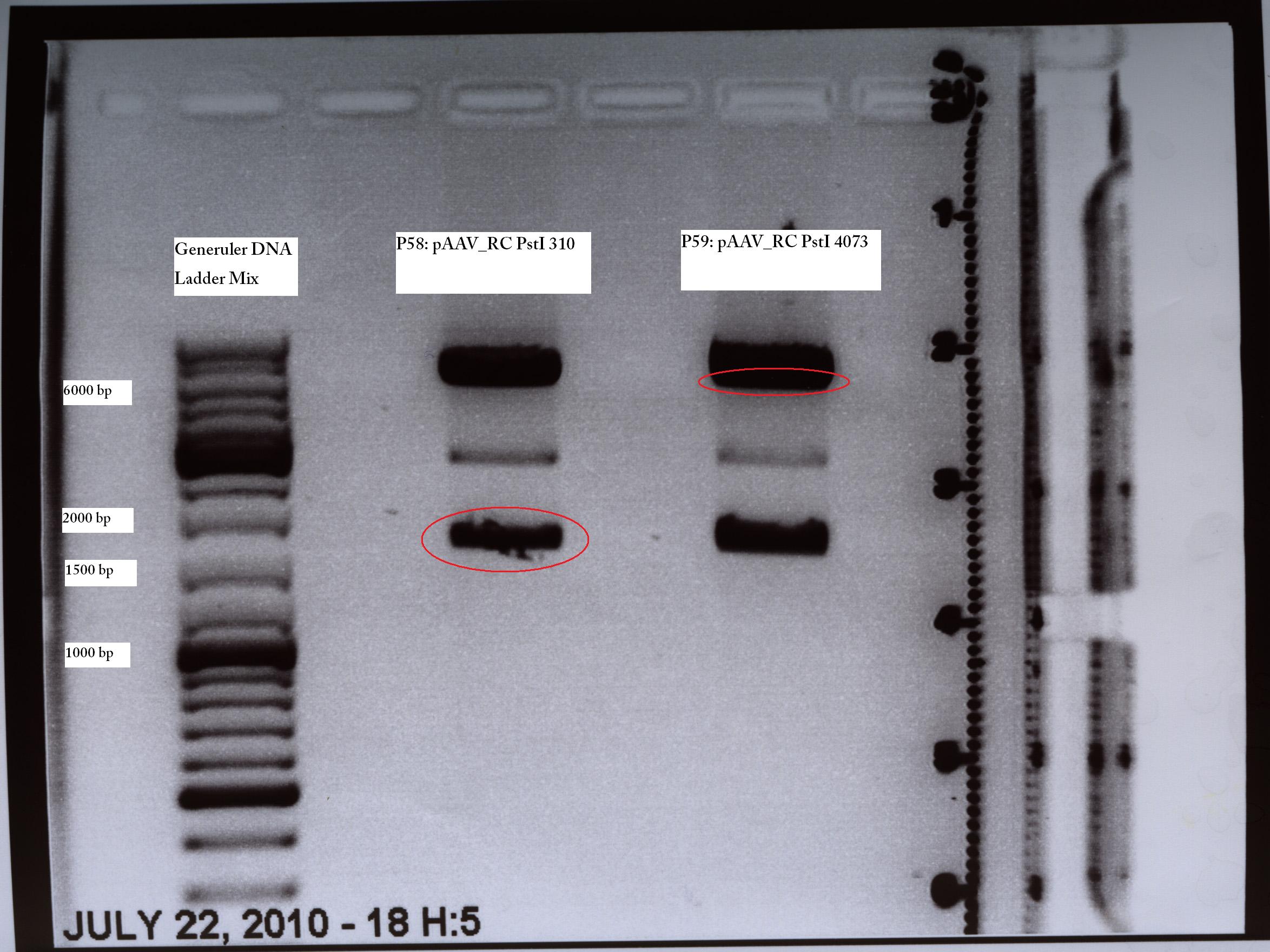
@Kira: ich habe die Clone benannt: SDM310 Nr. 1 ist jetzt P58 und sdm4073 nr 1 ist jetzt P59. Bitte eintragen.
TEST DIGESTION (PstI Quickchange Trafo from 13.07)
Investigator: Kira
FACS
Investigators: Kerstin, Anna
FACS analysis of different treated HT1080 cells was done (18 samples)
The first percentage refers to the living and fluorescent cells and the second percentage to the total amout of living cells.
The data are listed this way:
1. Plate I(A is up)
| A1 | A2 | A3 |
| B1 | B2 | B3 |
1= Plate 1 A1 : 0% YFP-positive Cells (there were only a few cells in this sample)
2= Plate 1 A2 : 27,7% YFP-positve Cells from 73,3%
3= Plate 1 A3 : 17,3% " from 63,1%
4= Plate 2 A1 : 32,9% " from 75,3%
5= Plate 2 A2 : 27,2% " from 76,5%
6= Plate 2 A3 : 30,7% " from 73,4%
7= Plate 3 A1 : 26,8% " from 76,0%
8= Plate 3 A2 : 30,5% " from 74,2%
9= Plate 3 A3 : 30,7% " from 75,2%
10= Plate 1 B1 : 6,25% " from 36,1%
11= Plate 1 B2 : 6,42% " from 68,4%
12= Plate 1 B3 : 0,05% " from 89,5%
13= Plate 2 B1 : 20,5% " from 72,5%
14= Plate 2 B2 : 15,7% " from 73,6%
15= Plate 2 B3 : 0,02% " from 85,4%
16= Plate 3 B1 : 19,5% " from 73,3%
17= Plate 3 B2 : 20,8% " from 75,7%
18= Plate 3 B3 : 0,07% " from 84,8%
The calculated value written into the table describes the fluorescent amount of cells from the living cells. Example: 1 A2 : 27,7% YFP-positve cells of 73,3% living cells : 27,7/73,3 = 0,378
1. Plate I(A is up) completely 10 µg
| 300µl dropped 0,00 | 300µl resuspended 0,378 | 450µl resuspended 0,274 |
| 600µl dropped 0,173 | 600µl resuspended 0,094 | control 5,6x10^-4 |
2. Plate II(A is up)completely 18 µg
| 300µl dropped 0,437 | 300µl resuspended 0,356 | 450µl resuspended 0,418 |
| 600µl dropped 0,283 | 600µl resuspended 2,1x10^-3 | control 2,3x10^-4 |
3. Plate III(A is up) completely 24 µg
| 300µl dropped 0,353 | 300µl resuspended 0,411 | 450µl resuspended 0,408 |
| 600µl dropped 0,266 | 600µl resuspended 0,275 | control 8,3x10^-4 |
Conclusions
- Higher amounts of Plasmids (24, 30µg) seems to have no obvious effect on YFP expression
- Theres no difference in YFP expression comparing resuspended samples to not resuspended samples
- The amount of dead HT cells is about 25%, it is not clear if it is either AAV induced or mechanic stress (Trypsin, washing procedure)
Next steps:
- does the amount of GOI (ng plasmids) has an effect on YFP expression
- compare transduced to non transduced cells (vitality), or in other words is the mechanic stress responsible for 25% dead cell ratio?
- comparison of transduction efficiency of different virus stock production routines
Plasmid Mini-Prep of pSB1C3_CFP
- experiment date: 15.07.2010 ; time:10.00
- name of investigator: Kerstin, Anissa
- new vector name:pSB1C3_CFP
Glycerol Stocks
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | |
| Bacteria strain | XL1 blue | XL1 blue | XL1 blue | XL1 blue |
| Plasmidname | pSB1C3_CFP | pSB1C3_CFP | pSB1C3_CFP | pSB1C3_CFP |
| Date | 15.7.2010 | 15.7.2010 | 15.7.2010 | 15.7.2010 |
| given glycerol-stock no. | B38 | B39 | B40 | B41 |
| given plasmid no. | p48 | p51 | p52 | p53 |
Test digestion
- buffer used: 3 ; Restriction-enzymes used: Enzyme 1 (no. Lab:48)PstI ; Enzyme 2 (no.Lab:23) EcoRI
- Plasmid
- Given Plasmid-Number: p48; DNA concentration: 195,1 ng/µl ;
- Given Plasmid-Number: p51; DNA concentration: 230,1 ng/µl;
- Given Plasmid-Number: p52; DNA concentration: 134,1 ng/µl;
- Given Plasmid-Number: p53; DNA concentration: 244,1 ng/µl;
Comments:p48 and p51 were sequenced
| Components | Volume/µL | Mastermix |
| DNA | Variable (800-1000 ng) | - |
| BSA (100x) | 1,5 | 7,5 |
| Buffer no.3 (10x) | 1,5 | 7,5 |
| Enzyme 1 (no. Lab:48 ) | 0,5 | 2,5 |
| Enzyme 2 (no. Lab:23 ) | 0,5 | 2,5 |
| H2O | variable | - |
| Total volume | 15 | - |
| Sample | Volume sample/ µl | Volume H2O / µl |
| p48 | 5 | 6 |
| p51 | 4 | 7 |
| p52 | 7 | 4 |
| p53 | 4 | 7 |
- Incubation: 1h
Agarose-Gel:
0,5 g Agarose, 50 TAE (1%), 3GELRED (3-6µl), at 110Volt, running time:30 minutes
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size (Geneious) |
|---|---|---|---|
| p48 | 15 µl | 3 µl | 700 bp |
| p51 | 15 µl | 3 µl | 700 bp |
| p52 | 15 µl | 3 µl | 700 bp |
| p53 | 15 µl | 3 µl | 700 bp |
- Marker: GeneRuler ladder mix
| Marker | Sample p48 /µl | Sample p51 /µl | Sample p52 /µl | Sample p53 /µl | |
|---|---|---|---|---|---|
| Lane | 8 µl | 18µl | 18µl | 18µl | 18µl |
Comments: Gel was leaky, therefore two samples "flew through". Plasmid p48 and p51 were sent for sequencing.
2nd try: First steps in producing ITR BioBricks via Fancy Method
Investigator: Hanna
Unfortunately it turned out, that the digestion step of the 1st try didn't work: After the transformation very much colonies grew on the plates. Sequencing delivered, that survival of the bacteria was due to re-ligated vector... Therefore the whole experiment will be performed again today.
Digestion of pGA14_Cerulean-CFP
1. Digestion with EcoRI and NotI:
| components | volume of vector /µl |
| DNA | 3.4 |
| BSA (10x) | 2 |
| Buffer 4 (10x) | 2 |
| Enzyme EcoRI-HF | 1 |
| Enzyme NotI-HF | 1 |
| H2O | 10.6 |
| Total volume | 20 |
Incubation: 1.5 hours, 37°C in thermocycler
Gel run: 20 µL sample + 4 µL loading dye (6x); 8µL marker
Expected fragment size: ~ 2900 bp
Gel extraction:
Sample weight: 0.31 g
Gel extraction was performed following the standard protocol. Abberation: After applying EB-buffer, the column was incubated for two minutes at 50°C.
DNA concentration: 43 ng/µL
1. Digestion with XbaI and PstI:
| components | volume of vector /µl |
| DNA | 3.4 |
| BSA (10x) | 2 |
| Buffer 4 (10x) | 2 |
| Enzyme XbaI | 1 |
| Enzyme PstI-HF | 1 |
| H2O | 10.6 |
| Total volume | 20 |
Incubation: 1.5 hours, 37°C in thermocycler
Gel run: 20 µL sample + 4 µL loading dye (6x); 8µL marker
Expected fragment size: ~ 2900 bp
Gel extraction:
Sample weight: 0.31 g
Gel extraction was performed following the standard protocol. Abberation: After applying EB-buffer, the column was incubated for two minutes at 50°C.
DNA concentration: 38.6 ng/µL
Digestion of pAAV_MCS with AlwNI
| components | volume of vector /µl |
| DNA | 17.86 = 5 µg |
| BSA (10x) | - |
| Buffer 4 (10x) | 3 |
| Enzyme AlwNI (no.Lab:120) | 3 |
| H2O | 6.14 |
| Total volume | 30 |
Incubation: 1.5 hours , 37°C in thermocycler
Gel run: 30 µL sample + 6 µL loading dye (6x); 8µL marker
Expected fragment sizes: 2982 bp = right ITR and 1674 bp = left ITR
Gel extraction:
- Sample: right ITR = 0.19 g
- Sample: left ITR = 0.17 g
Gel extraction was performed following the standard protocol. Abberation: After applying EB-buffer, the column was incubated for two minutes at 50°C. Further on 30 µL EB-buffer was used.
DNA concentration:
- right ITR: 59.8 ng/µL
- left ITR: 46.2 ng/µL :)
Digestion of ITR fragments with NotI and PstI
Digestion of right ITR fragment:
| components | volume of vector /µl |
| DNA | 30 µL |
| BSA (100x) | 0.5 |
| Buffer 4 (10x) | 3.74 |
| Enzyme NotI-HF | 1.5 |
| Enzyme PstI-HF | 1.5 |
| H2O | - |
| Total volume | 37.4 |
Incubation: 50 minutes, 37°C in thermocycler
Digestion of left ITR fragment:
| components | volume of vector /µl |
| DNA | 30 µL |
| BSA (100x) | 0.5 |
| Buffer 4 (10x) | 3.72 |
| Enzyme NotI-HF | 1.5 |
| Enzyme PstI-HF | 1.5 |
| H2O | - |
| Total volume | 37.2 |
Incubation: 50 minutes, 37°C in thermocycler
Gel run:
- due to the small fragment sizes, a 2% agarose gel was prepared.
- 5 µL loading dye was added to the whole volume of each sample; 8µL marker
- 115 V, 40 minutes
- Expected fragment sizes: ~ 150 bp (both)
- Problem: Small fragments, little amounts, blurred 150 bp bands -> fragments were cut out broad-minded.
Gel extraction:
- Sample: right ITR oben = 0.4 g
- Sample: left ITR = 0.36 g
Gel extraction was performed following standard protocol. After applying EB buffer to the columns, they were placed for 2 minutes into 50°C water bath and then centrifuged at 13200 rpm for 1 minute.
DNA concentration:
- right ITR: 4 ng/µL
- left ITR: 4 ng/µL
Hybridization of ITR-Präfix-Oligos
left ITR:
- 10 µL Oligo 1 (O43 = Präfix_for)
- 10 µL Oligo 2 (O44 = Präfix_rev)
- 4 µL 100 mM TrisHCl pH8
- 8 µL 5 mM MgCl2
- 8 µL H2O
Total: 40 mL
right ITR:
- 10 µL Oligo 1 (O47 = Präfix_for)
- 10 µL Oligo 2 (O48 = Präfix_rev)
- 4 µL 100 mM TrisHCl pH8
- 8 µL 5 mM MgCl2
- 8 µL H2O
Total: 40 mL
Because some of the oligos produce strong secondary structures, we used the ORIGAMI1 program:
1) initial denaturation: 99°C, 7 minutes
2) 99°C, 1 minute
3) 73 x repetition of 2) -> -1°C, R = 0.3 °C/sec
4) hold 4°C
Ligation of pGA14 + ITR + Präfix
1. right ITR
- Präfix: 2 µL
- ITR: 10 µL
- vector (pGA14): 5 µL
- buffer: 2 µL
- T4 ligase: 1 µL
Total volume: 20 µL
2. left ITR
- Präfix: 2 µL
- ITR: 10 µL
- vector (pGA14): 5 µL
- buffer: 2 µL
- T4 ligase: 1 µL
Total volume: 20 µL
3. control 1:
- vector 1 (pGA14, digested with EcoRI + NotI): 5 µL
- buffer: 2 µL
- T4 ligase: 1 µL
- H2O: 12 µL
Total volume: 20 µL
4. control 2:
- vector 2 (pGA14, digested with XbaI + PstI): 5 µL
- buffer: 2 µL
- T4 ligase: 1 µL
- H2O: 12 µL
Total volume: 20 µL
Samples will be incubating over night at 16°C.
To do: Trafo (Agar plates were already prepared.)
60.Labortag 16.07.2010
Continuation of Fancy ITR-method
Investigator: Hanna
Trafo
Trafo was performed following standard protocol. Aberration: Instead of 2 µL, 3 µL DNA was added to BL21 bacteria (50 µL).
right ITR
It turned out, that the strategy by digesting pGA14 with XbaI and PstI cannot work. pGA14 needs to be digested with EcoRI and PstI:
Digestion of pGA14_Cerulean-CFP with EcoRI and PstI
| components | volume of vector /µl |
| DNA | 3.4 |
| BSA (10x) | 2 |
| Buffer 4 (10x) | 2 |
| Enzyme EcoRI-HF | 1 |
| Enzyme PstI-HF | 1 |
| H2O | 10.6 |
| Total volume | 20 |
Incubation: 1 hour, 37°C in thermocycler
Gel run: 1% gel, 20 µL sample + 4 µL loading dye (6x); 8µL marker
Expected fragment size: ~ 2900 bp
Gel extraction:
Sample weight: 170 mg
Gel extraction was performed following the standard protocol. Abberation: After applying EB-buffer, the column was incubated for two minutes at 50°C.
DNA concentration: 26.8 ng/µL
Digestion of pAAV_MCS with NotI and PstI
In order to create ITRs with NotI and PstI restriction sites, the AlwNI-digestion step was skipped. Therefore 50% of the constructs will also contain the left ITR... this means: picking more colonies and sequencing!
| components | volume of vector /µl |
| DNA | 10.71 |
| BSA (10x) | 2 |
| Buffer 4 (10x) | 2 |
| Enzyme EcoRI-HF | 1 |
| Enzyme PstI-HF | 1 |
| H2O | 3.29 |
| Total volume | 20 |
Incubation: 1 hour, 37°C in thermocycler
Gel run: 2% gel, 20 µL sample + 4 µL loading dye (6x); 8µL marker
Expected fragment size: ~ 150 bp
Gel run delivered no satisfying results. Therefore the gel was discarded and the strategy changed: The residual right ITR solution of yesterday's elution will be taken for ligation.
Ligation of right ITR + Präfix + pGA14
- ITR: 7 µL
- pGA14: 7 µL
- Präfix: 1.5 µL
- Buffer: 2 µL
- T4 ligase: 1 µL
- H2O: 1.5 µL
Total: 20 µL
- pGA14: 7 µL
- Buffer: 2 µL
- T4 ligase: 1 µL
- H2O: 10 µL
Total: 20 µL
The ligation-solution will be incubating over night.
To do:
- Mini-Prep of pGA14_präfix_leftITR
- Trafo of pGA14_präfix_rightITR
- Midi-Prep of pGA14
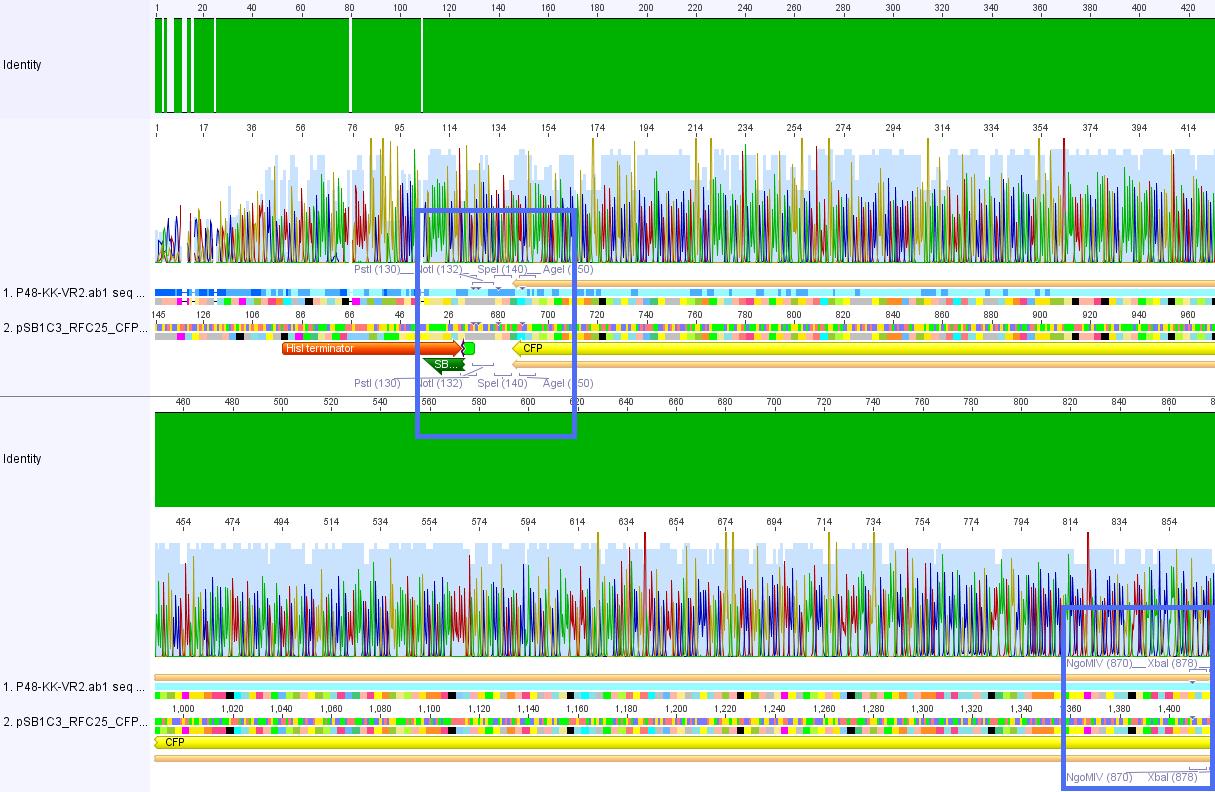
Results sequencing of pSB1C3_CFP
Investigator: Bea
Plasmid: pSB1C3_CFP
Primer used: VR2
Plasmid names: P48_KK and P51_AB
Comments: Sequencing results of cloning experiment performed at 14.-15.07.2010.
Results: Sequencing reads are good. Cloning of CFP into pSB1C3 can be confirmed. There are no insertions, transitions or deletions in the sequence or the iGEM restriction sites. Using this plasmid in order to create our iGEM BioBricks in RFC10 standard can be confirmed.
BioBrick production with hGH and beta-globin by cloning it into pSB1C3
Investigator: Bea
Plasmid: pSB1C3_CFP
Plasmid names: pSB1C3 (P51 or P48) and PCR prdouct of hGH and beta-globin
Comments: While cloning theoretically the hGH PCR product into the vector pSB1C3, it was recognized that the primers that were used for PCR for
- hGH RFC10 production and
- beta-globin intron RFC10 production
do not contain the required "G" at the end of the prefix and the "T" in the suffix add-on in the RFC10 standard. They were already used by performing a PCR with the add-on prefix and suffix primer, but we did not clone it into the pSB1C3 because of the wrong add-on tails. Therefore new primers were designed and ordered.
Ordered oligos:
hGH
- suffix (reverse)
- prefix (forward)
beta-globin
- suffix (reverse)
- prefix (forward)
CMV
- suffix (reverse)
- prefix (forward)
TK_GMK
- tk_gmk_prefix for
- tk_gmk_suffix_rev
- gmk_suffix_rev
To do: Upload cloning strategy of tk_gmk and BioBrick production
--Bea 15:21, 16 July 2010 (UTC)
Comment: reverse beta-globin primer is correct!? (Hanna)
Transfection
Investigator: Adrian
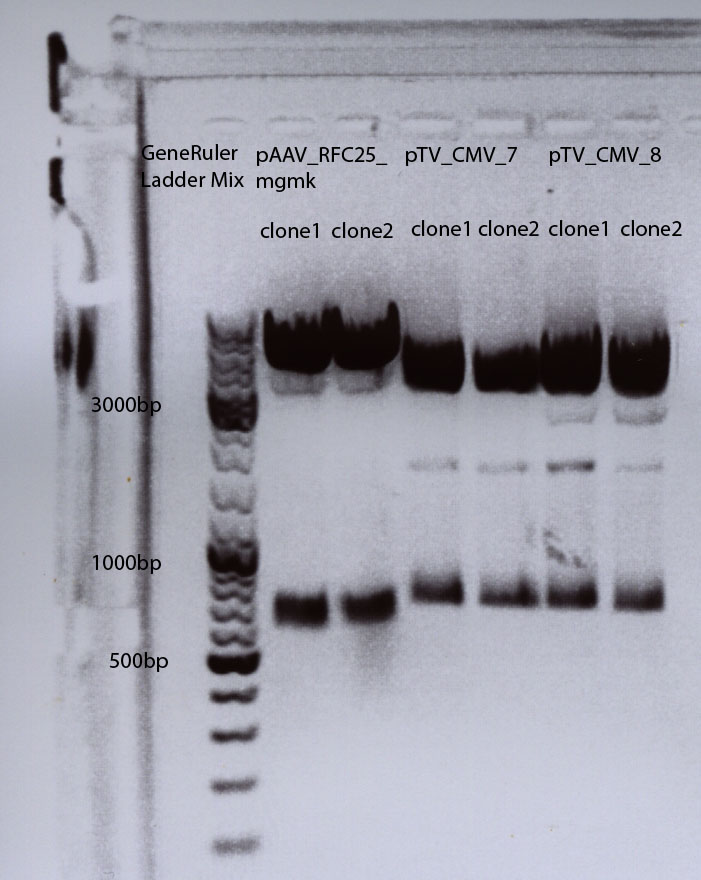
30µg Plasmid were used (pHelper + pAAV_RC+ pAAV_RFC25_mgmk_TK30)
VIRUS nix:pHelper + pAAV_RC
VIRUS 1 :pHelper + pAAV_RC+ pAAV_RFC25_mgmk_TK30 clone 1 : P54
VIRUS 2 :pHelper + pAAV_RC+ pAAV_RFC25_mgmk_TK30 clone 2 : P55
VIRUS 3 :pHelper + pAAV_RC+ pAAV_RFC25_mgmk_TK30 clone 3 : P56
Mini Prep of pAAV_RFC25_mgmk_TK30
Investigator: Adrian 4ml were preparated
- pAAV_RFC25_mgmk_TK30 clone1 conc: 291,43 ng/µl : P54
- pAAV_RFC25_mgmk_TK30 clone2 conc: 243,99 ng/µl : P55
- pAAV_RFC25_mgmk_TK30 clone3 conc: 247,00 ng/µl : P56
- pAAV_RFC25_mgmk_TK30 clone4 conc: 246,01 ng/µl : P57
Test digestion of all four plasmids to check if the mgmg_TK30 insert is present. Investigator: Patrick, Chris
Each digestion sample:
- 0,2 µl BSA (I picked the wrong tube. It should have been 2 µl BSA)
- 2 µl Buffer 4
- 0,5 µl XbaI
- 0,5 µl SpeI
- 13,8 µl H2O
- 3 µl DNA
Digestion: 65 minutes, 37°C. Results: No mgmk_TK30 insert detectable. I will try it again on saturday or sunday.
Preparation of 50x TAE and 0,5M EDTA
Investigators: Chris W., Patrick
1 L TAE (50x)
242 g tris base
57.1 ml glacial acetic acid
100 ml 0.5 M EDTA
ad 1 L Milipor
1 L EDTA (0.5 M)
146,13 g Ethylendiamintetraessigsäure
ad 800 ml Milipor
~20 g NaOH
pH mit NaOH auf 8 einstellen
ad 1 L Milipor
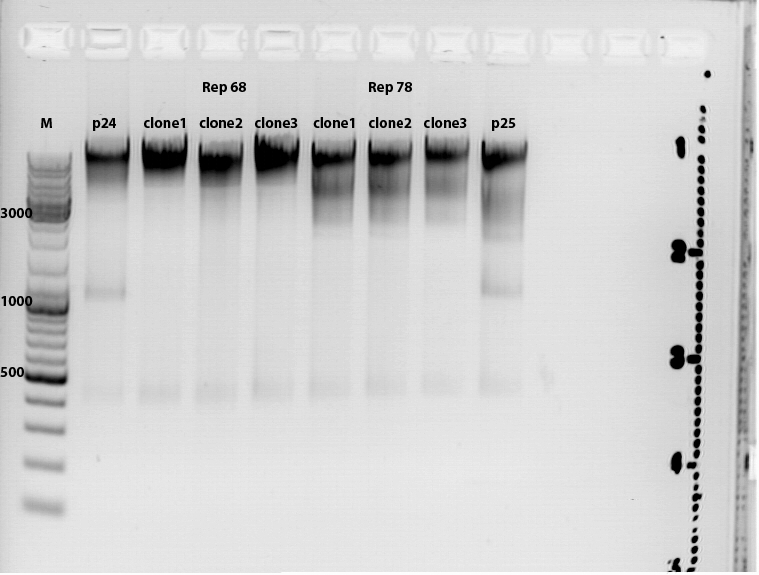
Test digestion of the QuickChange for PstI (310) and PstI (4073)
Investigator: Kira & Volker
The digestion showed two bands with apropirate lengt for each of the 6 clones as they were expected.
The constructs were sent to GATC for sequencing.
- pAAV_RC_SDM_PstI(310) with the primer GATC_std_QE-FP
- pAAV_RC_SDM_PstI (4073) with the primer GATC_std_M13_FP
Primers to convert the expression constructs into the iGEM standard were ordered
Investigator: Volker
- Praefix VP1ex
- Praefix VP2ex
- Praefix VP3ex
- Suffix VP123ex
- Praefix Rep68_78ex
- Praefix Rep 40_52ex
- Praefix AAPex
- Suffix Rep 40_68ex
- Suffix Rep 52_78ex
- Suffix AAPex
61.Labortag 17.07.2010:
Picking clones of pGA14_Präfix-leftITR Trafo
Investigator: Hanna
Bacteria also grew on control plates. Control means: Ligation of digested vector (pGA14_Cerulean) without insert (Präfix and left ITR). Therefore it seemed that somehow pGA14 religated (Can't be if vector digested completely). Therefore 10 colonies were picked, in order to increase the chance to get a successfully ligated plasmid.
Mini-Preps of pGA14_Präfix-lITR cultures
Investigator: Hanna
No glycerol stocks were prepared, because I'm going to perform the test digestion today.
10 Mini-Preps were performed following the standard protocol.
Test digestion of pGA14_Präfix-lITR (Ligation)
Investigator: Hanna
Test digestion
- buffer used: 4; Restriction-enzymes used: Enzyme 1: EcoRI-HF ; Enzyme 2: PstI-HF
- Plasmid: pGA14_Präfix_lITR 1 - 10 (all: ~ 400 ng/µL)
| Components | Volume/µL | Mastermix |
| DNA | 2.5 | - |
| BSA (10x) | - | 12 |
| Buffer 4 (10x) | - | 12 |
| Enzyme 1 EcoRI-HF | - | 6 |
| Enzyme 2 PstI-HF | - | 6 |
| H2O | - | 54 |
| Total volume | 10 | 90 |
- Incubation: 55 minutes
Agarose-Gel:
0.75 g Agarose, 50 ml TAE (1.5 %), 3 µL GELRED, at 115 Volt, running time: 30 minutes
Expected fragment size: Hopefully 160 bp = left ITR :) If pGA14_Cerulean religated the expected fragment size would be ~ 800 bp.
- Marker: GeneRuler ladder mix (6x)
| Marker | Sample 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 | Sample 6 | Sample 7 | Sample 8 | Sample 9 | Sample 10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lane | 8 µL | 10 µL | 10 µL | 10 µL | 10 µL | 10 µL | 10 µL | 10 µL | 10 µL | 10 µL | 10 µL |
Comments: Numbers refer to "colony 1, 2, 3,..."
Wonderful news: All 10 test digestions delivered positive results: 150 bp fragments and no 800 bp fragment was detectable. Means: left ITR + Präfix was inserted successfully into pGA14!!! :)
yees, thats great! Finally something worked with our ITRs. well done Hanna!! (Bea)
the most astonishingly test digestion gel of our entire working time! :)(Adrian)
Next step: Fusion of suffix to left ITR
Glycerol stocks of all 10 colonies were prepared: B46.1 - B46.10; Plasmid names: P62.1 - P62.10
Trafo of pGA14_Präfix_rITR
Investigator: Hanna
Transformation of BL21 bacteria with pGA14_Präfix_rITR which ligated over night was performed following standard protocol. Abberation: 3µL DNA was added to 50 µL cells.
To do: Mini-Prep + Test digestion of pGA14_Präfix_rITR; Sequencing of pGA14_Präfix_lITR and pGA14_Präfix_rITR.
62.Labortag 18.07.2010
Investigator Adrian:
Calculation of ganciclovir cymeven concentration
- Ganciclovir: m = 255,23 g/mol
- dissolved in PBS (?), NaCL (0,9%) ?
- Concentrations : 0,5 µM = 0,0000005M
- xg = (0,0000005 mol/l *255,23 g/mol)/(1l PBS resp. 1l 0,9% NaCl)
- x = 0,000128g
- x = 0,128 mg
- 0,128 mg solved in 1l of PBS = 1l of 0,5µM solution
- steril filtrated under steril bench
preparation of Aliquots:
Cellculture
The HEK-293 cells were split according to the standard protocol. The HT 1080 cells were split and sowed (3x10^5 cells each well). The amount of cells/ml was determined with the Neubauer-Meteringchamber: (8 cells counted/4)x2,2x20x10^4 = 888000 cells/ml. Two 6-well dishes could be prepared.
| only DMEM | 3x10^5 cells+DMEM | 3x10^5 cells+DMEM |
| 3x10^5 cells+DMEM | 3x10^5 cells+DMEM | 3x10^5 cells+DMEM |
Test digestion of pAAV_RFC_mgmk_tk30: P54, P55, P56, P57
Investigators: Chris, Patrick
To check whether the mutual mgmk_tk30 insert is present in the plasmid we digested it with AgeI and EcoRI. If the insert is present there will an DNA fragmet with a size of about 1700 bp.
| P54 | P55 | P56 | P57 |
| 291,4 ng/µl | 244 ng/µl | 247 ng/µl | 246 ng/µl |
Mastermix:
- 10 µl Buffer 4
- 4,4 µl AgeI
- 2,5 µl EcoRI
- no BSA (accroding to NEB)
- 83,1 µl H2O
- Total volume: 100 µl
17 µl of the mastermix were pipetted to 3 µl plasmid sample. Digestion time: 75 minutes.
Gelrun (1% Agarose), 55 minutes, 115 mV: Results: See picture. Interpretation of the results: ? The "upper" DNA seems to be undigested vector underneath the probably digested vector with an estimated size of about 5600 bp. The mutual insert (mgmk_tk30) should be about 1700 bp. There are probably several DNA coil structures visible but not our mutual insert. To do : ligate again.
Aferwards i noticed that i miscalculated the Mastermix.
63. Labortag 19.07.2010
rITR
Picking clones of pGA14_Präfix-rITR Trafo
Few bacteria also grew on control plates. Control means: Ligation of digested vector (pGA14_Cerulean) without insert (Präfix and right ITR).
6 colonies were picked, in order to increase the chance to get a successfully ligated plasmid.
Mini-Preps of pGA14_Präfix-lITR cultures
Investigators: Hanna, Jessy
Glycerol stocks were prepared: B47.1, B47.2, B47.3, B47.4, B47.6
6 Mini-Preps were performed following the standard protocol.
Test digestion of pGA14_Präfix-rITR (Ligation)
Investigator: Hanna, Jessy
Test digestion
- buffer used: 4; Restriction-enzymes used: Enzyme 1: EcoRI-HF ; Enzyme 2: PstI-HF
- Plasmid: pGA14_Präfix_rITR 1 - 6 (all: ~ 500 ng/µL) -> 1 µg
| Components | Volume/µL | Mastermix |
| DNA | 2 | - |
| BSA (10x) | - | 6.4 |
| Buffer 4 (10x) | - | 6.4 |
| Enzyme 1 EcoRI-HF | - | 4 |
| Enzyme 2 PstI-HF | - | 4 |
| H2O | - | 43.2 |
| Total volume | 10 | 64 |
- Incubation: 45 minutes
Agarose-Gel:
0.75 g Agarose, 50 ml TAE (1.5 %), 3 µL GELRED, at 90 - 100 Volt, running time: 45 minutes
Expected fragment size: Hopefully 160 bp = right ITR :) If pGA14_Cerulean religated the expected fragment size would be ~ 800 bp.
- Marker: GeneRuler ladder mix (6x)
| Marker | Sample 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 | Sample 6 | Marker | |
|---|---|---|---|---|---|---|---|---|
| Lane | 8 µL | 10 µL | 10 µL | 10 µL | 10 µL | 10 µL | 10 µL | 10 µL |
Comments: Numbers refer to "colony 1, 2, 3,..."
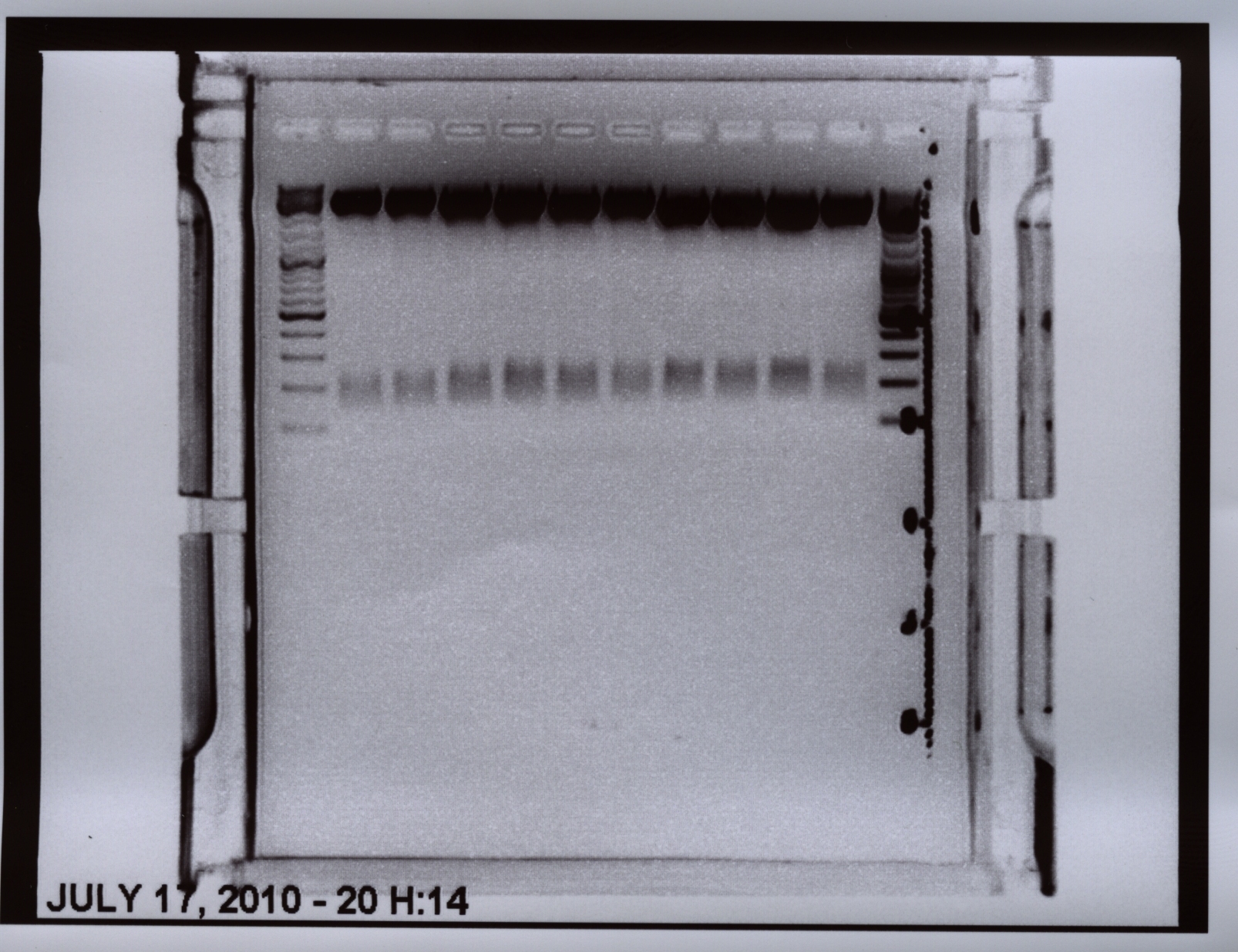
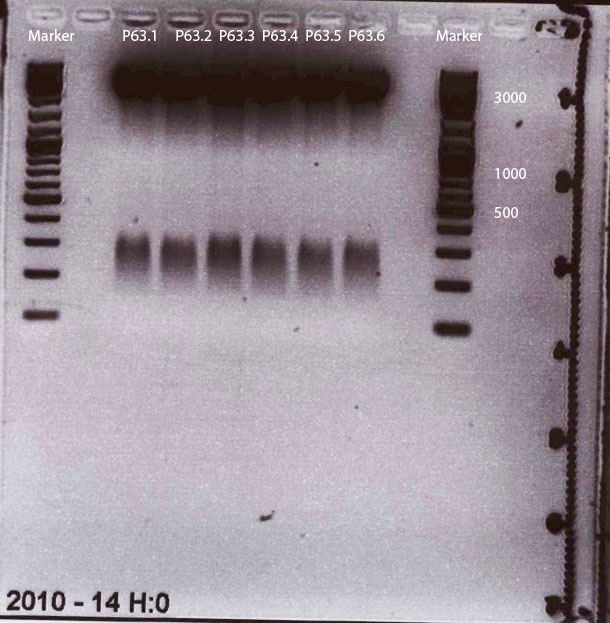
Wonderful news: All 6 test digestions delivered positive results: ~ 160 bp fragments and no 800 bp fragment were detectable. Means: right ITR + Präfix was inserted successfully into pGA14!!! :)
Next step: Ordering new oligos: Präfix of lITR, suffix of right and left ITRs, continuation of fancy cloning protocol
New Oligos (left ITR Präfix and Suffix + right ITR Suffix) were ordered.
Sequencing of ITRs
Investigator: Hanna
pGA14_Präfix_lITR and pGA14_Präfix_rITR were sent for sequencing to GATC:
1) pGA14_Präfix_lITR: c(P62.1) = 422.38 ng/µL -> V = 5 µL + 25 µL H2O
2) pGA14_Präfix_rITR: c(P63.1) = 501 ng/µL -> V = 4.2 µL + 25.8 µL H2O
Primer: GATC_std_pQE-FP
Quantitative real time PCR
Investigator: Hanna
Template: DNA of HT1080 cells transducted with 1) 10 µL and 2) with 30 µL virus stock solution.
Pipetting scheme:
Master Mix
- 2x QuantiFast SYBR Green PCR Mix: 400 µL
- Primer for and rev: 32 µL of 1:10 dilution = 10 µM
- Water: 208 µL
- Total: 640 µL
Template: 5 µL
To each template 20 µL master mix were added.
PCR program:
- Hold: 95°C, 5 minutes
- Cycling: 95°C for 10 seconds, 60°C for 30 seconds
- Melt: Ramp from 60°C to 95°C, rising by 1°C each step, wait for 45 seconds on first step, then wait for 5 seconds for each step afterwards. Aquire to Melt A on FAM.
Cloning strategy for gmk_tk added!!
Investigator: Bea
Media:Freiburg10 BioBrick GMK TK30 07 07 2010 with new oligos.pdf
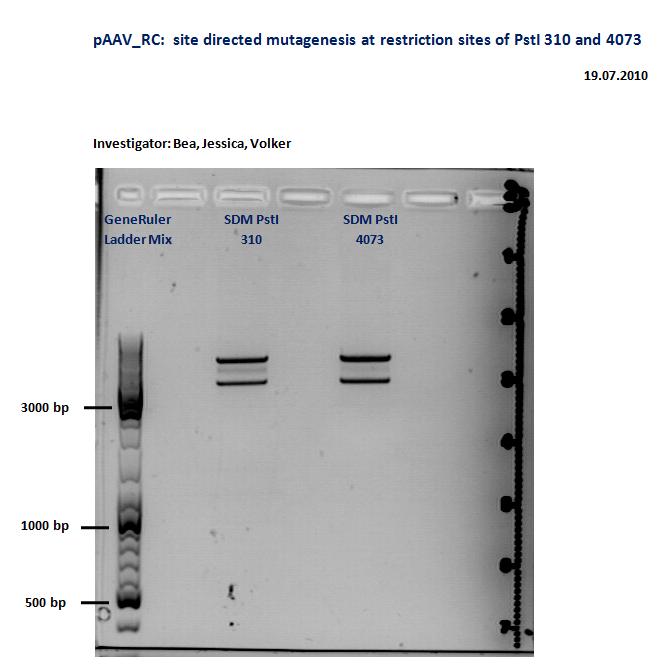
pAAV_RC site directed mutagenesis: Digestion of the two mutagenesis reaction of PstI site: 310 and 4073
Investigator: Volker
Digestion of pAAV_RC
| components | P58 /µl | P59 /µl |
| DNA | 4,34 | 3,76 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme XmnI (no.Lab: 66) | 1 | 1 |
| H2O | 10,66 | 11,24 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 |
Incubate at 37°C
- Gelextraction was prepared according to the protocol
- weight of gel: 310: 150 mg; 4073: 150mg
- weight of gel: 310: 150 mg; 4073: 150mg
- Nanodrop: 310: 5,5ng/µl; 4073: 7,0ng/µl
- Ligation was prepared according to the protocol
- Ratio: 310:4073 --> 2,52 µl:6,48µl
- Ratio: 310:4073 --> 2,52 µl:6,48µl
Ligation was frozen in -20°C
pHelper plasmid for Midi-Prep prepared
Investigator: Stefan
200ml DYT media + 200µl Amp were inoculated with pHelper-plasmid glycerol stock and put on shaker in 37°C room.
For Midi-Prep: Use Endotoxin-free Midi Prep not our Midi Prep!
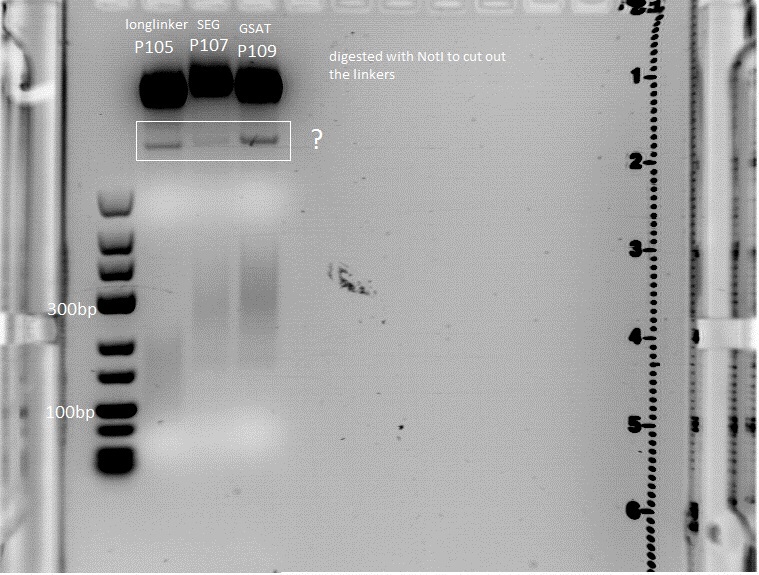
Trafo of IGEM 2009 linkers
- We borrowed several linkers/tags from Gerrit and did a Trafo in XL1B cells:
- small linker
- middle linker
- long linker
- Strep-Tag
- Split
- GSAT
- SEG
- Amount of DNA used: 0,5 µl
- Plates incubated at 37°C overnight
64. Labortag 20.07.2010
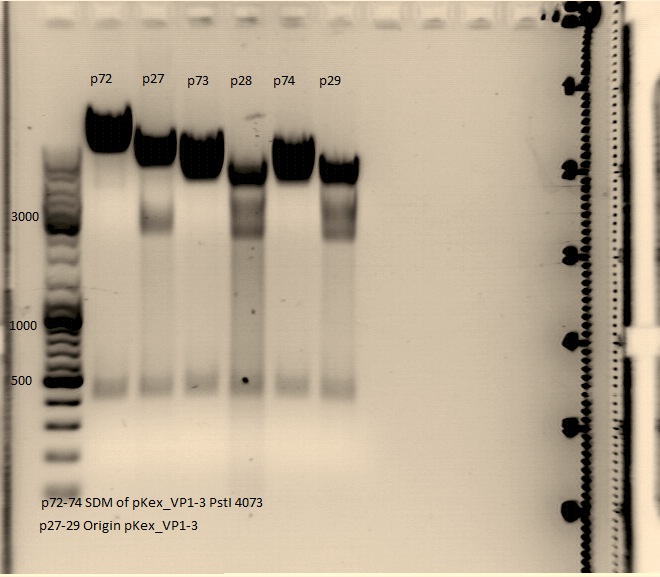
Quickchange site directed mutagenesis:
Investigators: Achim,Anissa
1) VP1ex P27.2
2) VP2ex P28.2
3) VP3ex P29.2
PCR reaction:
- 2.5 µL 10x Pfu Ultra II buffer
- 1) 1,44µl 2) 1,83µl 3) 1,76µl template (1:100 dilution)
- 0.34 µL primer SDM PstI (4073)forward (of 1:10 dilution)
- 0.35 µL primer SDM PstI (4073) reverse (of 1:10 dilution)
- 0,5 µL DMSO (primers form very strong secondary structures)
- 0.5 µL dNTP
- 1) 18,9 2) 18,5 3) 18,6 µL dH2O
- 0.5 µL PfuUltra II fusion (1.25 U)
-> end volume: 25 µL
PCR program:
Quickge
Trafo was carried out, using 2µl DNA, strain:XL1 blue
Transformation with ligation from july 19th
- used bacterial strain: BL21
- 2µL of DNA (pAAV_RC w/o PstI 310/4073)
- Trafo plate in 37°C room for incubating at 12.30
- no clones to pick at 22.00
Picking Clones from Linker Trafo
- clones were picked at 22.00
Endotoxin-free Midi Prep of pHelper
Investigators: Chris, Stefan
Midi-Prep was done according to protocol.
Result of the Midi-prep: 503,2 ng/µl
Plasmid (P64) was placed in Stratagene-Box.
Quantitative real time PCR
Investigator: Hanna
Because there was one positive non-template control (H2O) yesterday, we performed the qPCR one more time.
For this purpose we also calculated the plasmid pipetting for the standard curve again:
- c(P9) = 272 ng/µL --> 3 x 1:10 = 1:1000 dilution.
- 2.68 µL of the 1:1000 dilution was added to 52.32 µL H2O = 2600000 copies/µL.
- By performing 1:10 dilutions further 6 "standard samples" were created: 260000 copies/µL; 26000 copies/µL; 2600 copies/µL; 260 copies/µL, 26 copies/µL, 2.6 copies/µL
Template: DNA of HT1080 cells transducted with 1) 10 µL and 2) with 30 µL virus stock solution.
Pipetting scheme:
Master Mix
- 2x QuantiFast SYBR Green PCR Mix: 400 µL
- Primer for and rev: 32 µL of 1:10 dilution = 10 µM
- Water: 208 µL
- Total: 640 µL
Template: 5 µL
To each template 20 µL master mix were added.
Comment: To non-template control 2 17 µL + 3µL H2O was added (not enough master mix was left).
PCR program:
- Hold: 95°C, 5 minutes
- Cycling: 95°C for 10 seconds, 60°C for 30 seconds
- Melt: Ramp from 60°C to 95°C, rising by 1°C each step, wait for 45 seconds on first step, then wait for 5 seconds for each step afterwards. Aquire to Melt A on FAM.
Cloning of p51(pSB1C3_RFC25) for His-tag insert
Investigators: Anna, Stefan
| components | V (p51)/ µl |
| DNA | 5 |
| BSA (10x) | 2 |
| Buffer 4 (10x) | 2 |
| Enzyme: SpeI (no.Lab:71) | 1 |
| Enzyme: NgOMIV (no.Lab: 113) | 1 |
| H2O | 9 |
| Total volume | 20 |
1% Agarose-gel, 3µl Gelred
Loading dye(5x): 4µl
Loading plan: marker (10µl), free, sample (24µl)
expected size of fragment: 2080 bp
65. Labortag 21.07.2010
Sequencing of ITRs
Investigator: Hanna
P62.1 (=left ITR + RFC23-Präfix in pGA14) and P63.1 (= right ITR-Präfix in pGA14) were sequenced:
Sequencing delivered, that the präfixes were fused successfully to the ITRs. The mismatch (orange box) was due to the alignment with the RFC10 präfix... (oligos were ordered!), what means, that the sequencing is OK.
Interestingly it seems, that not only we had problems with PCR of the ITR -> Very strong secondary structures. Even sequencing of the ITRs seems impossible under normal conditions: Exactly at the point where the ITR sequence starts the sequencing signal delivered no clear results.
Conclusion: Fancy method worked until now.
Quantitative real time PCR
Investigator: Hanna
Because the non-template control and the not-transduced cell-DNA controls delivered positive PCR results for the CMV-promoter, a control qPCR was prepared:
1. not-transduced cell-DNA controls (K1 and K2) were diluted: 1:100
2. Oligos (O20, O21) were 1:10 diluted
3. Master Mix was prepared:
- 75 µL SYBR Green
- 39 µL H2O
- 6 µL Oligos
4. Pipetting scheme:
- 1. K1
- 2. K2
- 3. NTC1
- 4. NTC2
- 5. Sdt7
Plasmid Mini-Prep
- experiment date: 21.07.2010 ; time: 10:30Uhr
- name of investigator: Anissa, Kerstin
Glycerol Stocks
| Clone 1 | Clone 2 | Clone 3 | Clone 4 | Clone 5 | Clone 6 | Clone 7 | |
| Bacteria strain | XL1 B | XL1 B | XL1 B | XL1 B | XL1 B | XL1 B | XL1 B |
| Plasmidname | pGA14_middle linker | pGA14_GSAT linker | pGA14_long linker | pGA14_split | pGA14_strep | pGA14_short linker | pGA14_SEG |
| given glycerol-stock no. | B48 | B49 | B50 | B51 | B52 | B53 | B54 |
| given plasmid no. | p65 | p66 | p67 | p68 | p69 | p70 | p71 |
Comments: Different Linker-fragments are very short (5-178bp) --> no Test-digestion; directly sequencing
Results of Trafo of cloning pAAV_RC SDM of PstI 310 and 4073
Investigator: Bea
Comments: Cloning together of the two site-directed mutagenesis performed with the pAAV_RC
Results: no clones grown on LB-agar plate, because pAAV_RC was digested with only one blunt-end restriction enzyme. Luckily, th enzyme vut twice, and one time in the Amp gene. Therefore wrong ligation of the two fragments lacked the Amp resistance.
To do: new cloning together of pAAV_RC PstI 310 and 4073 with TWO other enzymes.
PCR with pUB_V5_mGMK_TK30
Investigator: Adrian, Bea
Comments: Perform PCR with two primers
- pmGMK_TK30_prefix_RFC25_for
- pmGMK_TK30_suffix_RFC25_rev
and pUB_V5_hisB_mGMK_tk30 (P15) and clone it into pAAV_RFC25.
PCR:
- PCR program: GMKTK
| components | volume in µl |
| 5x Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| pMGMK_TK30_RFC25_prefix_for (060) (1:10 dilution) | 2,5 (0,5 µM) |
| pMGMK_TK30_RFC25_suffix_rev (063) (1:10 dilution) | 2,5 (0,5 µM) |
| DNA template: pUB_V5_hisB_mgmk_tk30 | ~2 µL of 1:1000 dilution |
| DMSO | - |
| Phusion polymerase | 0,5 |
| H2O | 31,5µL |
| Total volume (e.g. 50 µl) | 50 |
Comments:New Oligos (pmGMK_TK30_prefix_RFC25_for and pmGMK_TK30_suffix_RFC25_rev) with add-on tails (prefix and suffix), but TK30 still contains three iGEM restriction sites but will be ready for use in cell culture
Result of PCR product (theoretically)
Digestion of pAAV_RFC25
- Vector: name:pAAV_RFC25 number:p42
- new vector name: pAAV_RFC25_MGMK_TK30
- buffer used:4 ; Restriction-enzymes used: Enzyme 1 (no. Lab:149) AgeI ; Enzyme 2 (no.Lab:63) XbaI
- DNA concentration (vector):321,5 ng/µl
Comments: Digestion of pAAV_RFC25 wihle performing PCR with P42
| components | volume of vector /µl | volume of insert (=PCR product) /µl |
| DNA | 4,73 | - |
| BSA (100x) | 2 | - |
| Buffer 4 (10x) | 2 | - |
| AgeI (no.Lab:149) | 1 | - |
| XbaI(no.Lab:63) | 1 | - |
| H2O | 9,27 | - |
| Total volume (e.g. 15,20,25,30 µl) | 20 | - |
- Incubation: Vector: 1.5 h
Agarose-Gel:
- 0,5 g Agarose
- 50 ml TAE (1%)
- 3 µl GELRED (3-6µl)
- 110 Volt, running time: 50 minutes
Results of digestion with XbaI and AgeI of pAAV_RFC25 and PCR with pUB_V5_gmk_tk30
Result: Gel was loaded with 50µL of pCR product and 20µL of pAAV_RFC25 digestion. Expected bands can be seen, but pAAV_RFC25 too high and there are some additional bands. We cut out only the intensive band at around 1700 bp.
Comment:After a PCR there can be two ways to proceed with the PCR product:
- load 7-8µL of PCR sample on an analytical gel and watch agarose gel for additional bands. If there are some more bands than expected load sample on preparative gel.If there aren´t any additional bands. proceed with digestion of PCR product in order to ligate it into vector.
- load total PCR sample on preparative gel. Ask Sven or Tobi in which volume you should elute after cutting out the gel (Gel extraction), but you should elute sample at least in 30µL.
- After gel extraction was performed, concentration of the two constructs was measured and revealed:
- c(pAAV_RFC25)= 27,33ng/µL
- c(gmk_tk30)= 24,53ng/µL
- After the gel extraction, the PCR product gmk_tk30 was digested again with XbaI and AgeI
- and purified with the PCR purification Kit provided by Qiagen
Digestion of the PCR product gmk_tk30
| components | volume of vector /µl | volume of insert (=PCR product) /µl |
| DNA | - | 30 |
| BSA (100x) | - | 0,5 |
| Buffer 4 (10x) | - | 4 |
| AgeI (no.Lab:149) | - | 1 |
| XbaI(no.Lab:63) | - | 1 |
| H2O | - | 3,5 |
| Total volume (e.g. 15,20,25,30 µl) | - | 40 |
- Incubation: 1:45 h
- Ligation and Trafo were performed following standard protocols.
Gelextraction, Ligation and Transformation for cloning of pSB1C3_His_RFC25 (continuation of cloning from 64th lab day)
Investigator: Bea, Kerstin, Stefan
Extraction
- Gel extraction performed following standard protocol provided by Qiagen
Ligation
- Measure DNA-concentration with Nanodrop
- c(pSB1C3) = 18,2,8 ng/µL
- c(His-tag) = 11,2 ng/µL
- Calculation of volume needed for ligation:
- c(pSB1C3) = 8,26 µL
- c(His-tag) = 0,74 µL
Transformation
- Transformation has been followed the standard protocol Media:Freiburg10_Cloning Protocol.pdf
Deviation of standard protocol added: medium added to cells before incubation on thermomixer changed to DYT. Standard protocol changed.
Picking clones pf VP1-3 pEX plasmids from DKFZ
Investigator: Bea
Two clones of each plasmid were picked and inoculated in 10mL DYT medium containing 10 µL ampicillin
- VPex-1 clone 1
- VPex-1 clone 2
- VPex-2 clone 1
- VPex-2 clone 2
- VPex-3 clone 1
- VPex-3 clone 2
and they were put in the 37°c room at 19:15
66. Labortag 22.7.2010
cell culture
HT 1080 cells were split according to the standart protocol. Investigator: Adrian
Mini Prep and test digestion pKEX_VP1-3 w/o PstI(4073)
Investigators: Kerstin, Jessica
PstI should be deleted in VP1, VP2, VP3
- Glycerol stock were prepared:
- B55-B60
- Plasmids
- pKEX_VP1 clone1:488,2 P72
- pKEX_VP2 clone1:484,8 P73
- pKEX_VP3 clone1:485,8 P74
- pKEX_VP1 clone2:499,2 P75
- pKEX_VP2 clone2:511,7 P76
- pKEX_VP3 clone2:470,4 P77
Test digestion of three plasmids to check if the PstI 4073 is deleted (compared to the origin P27-P29)
| components | P72 /µl | P27 /µl | P73/µl | P28/µl | P74/µl | P29 |
| DNA | 1,6 | 1,7 | 1,6 | 8,1 | 7,5 | 7,4 |
| BSA (10x) | 1 | 1 | 1 | 1 | 1 | 1 |
| Buffer 4 (10x) | 1 | 1 | 1 | 1 | 1 | 0,5 |
| H2O | 10,9 | 10,8 | 10,9 | 4,4 | 5,0 | 5,1 |
| Total volume (e.g. 15,20,25,30 µl) | 15 | 15 | 15 | 15 | 15 | 15 |
Digestion: 45 minutes, 37°C. Results:
Deletion was succesful, there are still 2 PstI restriction sites in P72-74
Cloning together of pAAV_RC PstI 310 and 4073 with two enzymes
Clone P58 (pAAV_RC_SDM310.1) and P59 (pAAV_RC_SDM4073) will be digested with HindII and SpeI. The smaller fragment (about 1700 bp) of P58 and the larger fragment of P59 (about 5900 bp) will be ligated to receive a plasmid carrying both mutations.
| components | volume of vector /µl (P59) | volume of insert /µl (P58) |
| DNA | 2 | 5,5 |
| BSA (10x) | 2 | 2 |
| Buffer 2 (10x) | 2 | 2 |
| HindIII | 2 | 2 |
| SpeI | 2 | 2 |
| H2O | 12 | 9,5 |
| Total volume | 20 | 20 µl |
Incubation: at 37°C, 70 minutes.
The Gelextraction was performed according to the standard protocol.
Weight of P58 gel extract: 50 µg
Weight of P59 gel extract: 98 µg
The received concentrations (measured by nanodrop):
P58: 3 ng/µl , P59: 4,7 ng/µl
Quickligation was not performed according to the standard protocol:
- 2 µl P58 Insert
- 2 µl P59 Vector
- 5 µl Quickligase buffer (2x)
- 1 µl Quickligase
- Total volume: 10 µl
Incubation: 15 minutes at roomtemperature.
Transformation was performed according to the standard protocol with E. coli BL21. The agar plates were put into the 37°C room at 23:00.
New Ampicillin was prepared and put into the -20°C freezer. Investigators: Chris, Patrick
Sequencing of linker
Investigator: Anissa
| Linker | Sequencing |
| p65: middle linker | was sucessful |
| p66: GSAT linker | sequence didn’t arrive yet |
| p67: long linker | was sucessful |
| p68: split linker | false, no linker but YFP.Gerrit’s searching for the right sample |
| p69: strep linker | the suffix of strep has a double AgeI! That’s why we have to cut off next time with AgeI (and digest long enough) |
| p70: short linker | was sucessful |
| p71: SEG linker | was sucessful |
middle linker:
GSAT linker:
long linker:
Split linker:
Strep tag:
Short linker:
SEG linker:
Biobricks of hgH and ß-Globin
Investigators: Stefan, Anna
Could you upload some agarose gels eg the gels after the digestion and the PCR? Thanks a lot! (Bea)
Comments: Perform PCR with two primers
PCR of ß-Globin and hgH:
- DNA template: pAAV_MCS; concentration: 360 ng/µl
- PCR program: BGLOB
| components | volume in µl |
| 5x Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| ß_globin_suffix (085) (1:10 dilution) | 2,5 (0,5 µM) |
| ß_globin_prefix (086) (1:10 dilution) | 2,5 (0,5 µM) |
| DNA template: pAAV_MCS (1:1000) | 5,6 µL of 1:1000 dilution |
| DMSO | - |
| Phusion polymerase | 0,5 |
| H2O | 27,9 µL |
| Total volume (e.g. 50 µl) | 50 |
- PCR program: HGH
| components | volume in µl |
| 5x Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| (083) (1:10 dilution) | 2,5 (0,5 µM) |
| (084) (1:10 dilution) | 2,5 (0,5 µM) |
| DNA template: pAAV_MCS (1:1000) | 5,6 µL of 1:1000 dilution |
| DMSO | - |
| Phusion polymerase | 0,5 |
| H2O | 27,9 µL |
| Total volume (e.g. 50 µl) | 50 |
Digestion of pSB1C3_RCF25_CFP:
- DNA-Concentration: 230,9 ng/µl
| components | volume of vector /µl |
| DNA | 5 |
| BSA (10x) | 3 |
| Buffer 4 (10x) | 3 |
| PstI | 1 |
| XbaI | 1 |
| H2O | 17 |
| Total volume (e.g. 15,20,25,30 µl) | 30 |
Gel running of digested vector and PCR products
- 1% Agarose gel
- 3 µl Gelred, 8 µl DNA-Ladder-Mix
- 125 Volt, running time: 40 minutes
| Sample/µl | Loading dye (5x)/µl | Expected size/bp |
| Vector: 30 | 6 | 2080 |
| ß-Globin: 50 | 10 | 524 |
| hgH: 50 | 10 | 479 |
Gelextraction
- Gel extraction performed following standard protocol provided by Qiagen
| Sample/ | Weight/g | QG buffer/µl |
| Vector | 0,13 | 390 |
| ß-Globin | 0,28 | 840 |
| hgH | 0,24 | 720 |
Digestion of PCR products
| components | volume of vector /µl |
| DNA | 30 |
| BSA (100x) | 0,5 |
| Buffer 4 (10x) | 4 |
| PstI | 1 |
| XbaI | 1 |
| H2O | 3,5 |
| Total volume (e.g. 15,20,25,30 µl) | 40 |
Purification of PCR products, Ligation, Transformation
procedure has been followed a different protocol (see Sven); has to be added in the standard protocols !
- Measure DNA-concentration with Nanodrop:
c(pSB1C3) = 12,42 ng/µL
c(ß-Globin) = 51,93 ng/µL
c(hgH)= 36,87 ng/µL
- Calculation of volume needed for ligation:
c(pSB1C3) = 7,3 µL
c(ß-Globin) = 1,7 µL
c(pSB1C3) = 7,62 µL
c(hgH) = 1,38
Note: after heatshock cells were put on the shaker without adding 750 µl DYT, it was recognized and added later after 25 min of shaking !
Picking clones of pSB1C3_His_RFC25 and pAAV_RFC25_GMK_TK30 for Mini-Prep
Investigators: Stefan
Three clones of each construct were picked and inoculated in 10mL DYT medium containing 10 µL ampicillin (pAAV_RFC25_GMK_TK30) or 10 µL chloramphenicol (pSB1C3_His_RFC25).
They were put in the 37°C room at 20:33.
67. Labortag 23.7.2010
Picking clones of pAAV_RC_1.1
Investigator: ChrisW, Patrick
The inoculated agar plates from 'Cloning together of pAAV_RC PstI 310 and 4073 with two enzymes' were checked for colonies. Three clones were picked and in each case inoculated 20 ml DYT + Amp at 10:20.
Mini prep of two pAAV_RC_SDM_PstI_310_4073. Results: It was checked visually whether enough E. coli BL21 cells had grown. Therefore only two midi preps were carried out, according to the standard protocol (Final elution with 50 µl). Unfortunately both of them yielded a quite low plasmid concentration, probably due to the short growth time (7 h).
pAAV_RC mini pstI 301,4073: 44,7 ng/µl
pAAV_RC mini 2 pstI 301,4073: 83,1 ng/µl
30 µl of both plasmids were sent for sequencing to GATC, named PS1 and PS2. Primers used: GATC_std_PolyC-D and GATC_std_SK.
Glycerol stock:
B 67: pAAV_RC_SDM_PstI_310_4073 (1)
B 68: pAAV_RC_SDM_PstI_310_4073 (2)
Results of Trafo Biobricks of hgH and ß-Globin
Investigator: Anissa, Bea
Comments: No clones of pSB1C3_hGH and pSB1C3_betaGlobin grown on trafo plate. Possible reason can be the forgotten DYT medium added to the cells after the heat-shock. Cells were incubated at 37°C for 40 minutes without medium and were added afterewards.
TO DO: Repeat ligation of
- hgh PCR product and
- beta-globin PCR product
- Measure DNA-concentration with Nanodrop:
c(pSB1C3) = 38,9 ng/µL
c(ß-Globin) = 51,93 ng/µL
c(hgH)= 36,87 ng/µL
- Calculation of volume needed for ligation:
c(pSB1C3) = 3,8 µL
c(ß-Globin) = 5,2 µL
c(pSB1C3) = 3,8 µL
c(hgH) = 5,2
Attention: There was not enough cut vector. In both Ligations were only 2,2 µl of vector added. So incubation lasted 15 minutes and for transformation 4 µl were used.
Mini-Prep and Test digestion of pAAV_RFC25_mGMK_TK30
Investigator: Adrian, Bea
Comments: Mini-Prep, test digestion and sequencing is carried out in order to verify insertion of the PCR product mGMK_TK30 with RFC25 add-on tails into pAAV_RFC25(overhangs). Aim is to use the construct in order to conduct experiments with the gmk_tk30 fusionenzyme.
Mini-Prep of pAAV_RFC25_mGMK_TK30 clone 1 to 3: After performing Mini-Preps of clone 1 - 3 (elution volume with EB buffer was 50 µL) , concentration was measured and revealed following concentrations:
- P81 = pAAV_RFC25_mGMK_TK30 clone 1: c = 486 ng/µL
- P82 = pAAV_RFC25_mGMK_TK30 clone 2: c = 426 ng/µL
- P83 = pAAV_RFC25_mGMK_TK30 clone 3. c = 457 ng/µL
Test digestion of pAAV_RFC25_mGMK_TK30 clone 1 to 3:
- P81 = pAAV_RFC25_mGMK_TK30 clone 1
- P82 = pAAV_RFC25_mGMK_TK30 clone 2
- P83 = pAAV_RFC25_mGMK_TK30 clone 3
| components | volume of P81/µl | volume of P82/µL | volume of P83/µL |
| DNA | 2 | 2 | 2 |
| BSA (100x) | 1,5 | 1,5 | 1,5 |
| Buffer 4 (10x) | 1,5 | 1,5 | 1,5 |
| PstI | 0,5 | 0,5 | 0,5 |
| XbaI | 0,5 | 0,5 | 0,5 |
| H2O | 9 | 9 | 9 |
| Total volume /µl | 15 | 15 | 15 |
Expected sizes:
- mGMK_TK30: 1700bp
- pAAV_RFC25: 4600bp
Results of samples loaded 1% agarose gel and ran 50 minutes at 110 V:
Sequencing of pAAV_RFC25_mGMK_TK30
70ng/µL were sent to GATC from each clone. They were labeled:
- AF_P81 (clone 1)
- AF_P82 (clone 2)
- AF_P83 (clone 3)
Mini-Prep of pSB1C3_His6
Investigator: Adrian
pSB1C3_6xHis clone 1 = P84 : 107,8 ng/µl (eluted with 50µl)
pSB1C3_6xHis clone 2 = P85 : 122,8 ng/µl (eluted with 50µl)
pSB1C3_6xHis clone 3 = P86 : 98,8 ng/µl (eluted with 50µl)
68. Labortag 24.7.2010
Cell culture: Seeding of HEK293 for Transfektion with P81, P82 and P83
Investigator: Adrian
- unfortunately the T75 flask with HEK-cells was overpopulated => to much cells =>yellow medium and differentiation of the HEK-cells => thrown away
- I had to thaw cells, (P4) so there'll be no transfection at Monday :(
Cell culture: Transduction of HT1080 with AAVs
Investigator: Adrian
I. Plate I(A is up)
| control: no virus added | virus without GOI added | virus with 10µg amount of GOI added |
| control: no virus added | virus without GOI added | virus with 10µg amount of GOI added |
II. Plate II(A is up)
| control: no virus added | virus without GOI added | virus with 10µg amount of GOI added |
| control: no virus added | virus without GOI added | virus with 10µg amount of GOI added |
III. Plate II(A is up)
| control: no virus added | virus without GOI added | virus with 10µg amount of GOI added |
| control: no virus added | virus without GOI added | virus with 10µg amount of GOI added |
Discussion round nr. 4
Investigators: Adrian and his evil twins
- adrian created a poster
Stocktaking of superior goods
Investigator: Adrian
superior goods:
- DYT/LB medium
- tips
- LB-agar
69. Labortag 25.7.2010
Clone picking from pSB1C3_hgH and pSB1C3_betaglobin
Investigator: Bea
Comments: Clones were very small and not many clones could be detected. Possible reasons:
1) The ratio for a proper ligation need to be 3:1 insert: vector. There was not enough plasmid of the digested vector pSB1C3.
2) The digested fragments of the PCR products hGH and beta-globin and the cutted vector were stored at -20°C. This could lead to the bad results of the transformation, because the overhangs were degraded.
Two clones were picked of each trafo plate:
- pSB1C3_hGH
- pSB1C3_betaglobin
They were inoculated in 10mL DYT medium containing 10µL chloramphenicol and put into 37°C room for incubating. The reason for picking only two clones was that not many more clones could be detected easily.
70. Labortag 26.7.2010
Mini Prep and test digestion pSB1C3_hgh and pSB1C3_beta-globin
Investigators: Anissa, Kerstin
- Glycerol stock were prepared:
- B69-B71
- Plasmids
- pSB1C3_hgh clone1: 139 ng/µl p89
- pSB1C3_hgh clone2: 209,4 ng/µl p90
- pSB1C3_beta-globin clone1: 217,5 ng/µl p91
Incubation time: 1,5h Incubation temperature:37°C
Test digestion with Xba and PstI to check if hgh and beta-globin were cloned succesfully into the pSB1C3, respectively.
| components | p89 /µl | p90 /µl | p91/µl |
| DNA | 4,3 | 4,3 | 4,1 |
| BSA (10x) | 1,5 | 1,5 | 1,5 |
| Buffer 4 (10x) | 1,5 | 1,5 | 1,5 |
| H2O | 6,7 | 6,7 | 6,9 |
| Total volume (e.g. 15,20,25,30 µl) | 15 | 15 | 15 |
Test digestion P87: pAAV_RC_PstI-301-4073 clone 1 and P88: pAAV_RC_PstI-301-4073 clone 2
Investigators: Chris W.
- Plasmids
- pAAV_RC_PstI-301-4073 clone1: 44,2 ng/µl p87
- pAAV_RC_PstI-301-4073 clone2: 83,2 ng/µl p88
- pAAV_RC: 429,8 ng/µl p50
Incubation time: 1,5h Incubation temperature:37°C
Test digestion with PstI 0,5µl
| components | p87 /µl | p88 /µl | p50/µl |
| DNA | 13,6 | 7,2 | 0,72 |
| BSA (10x) | 1,5 | 1,5 | 1,5 |
| Buffer 4 (10x) | 1,5 | 1,5 | 1,5 |
| H2O | 2,9 | 9,3 | 15,78 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 | 20 |
Oligos for hTERT promoter
Investigator: Bea
Oligos were ordered to modify hTERt promoter in order to obtain hTERT in the iGEM RFC10 standard.
Retrafo with pTVvector containing the CMV promoter
Investigator: Bea
Comments: In order to obtain the CMV promoter in the correct RFC10 standard, oligos werde designed and ordered. For using the plasmid obtained from the iGEM team 2008, a retrafo has to be performed.
Re-transformation:
- Construct used: expressionvector + CMV (BBa_TV_CMV)
- clone 7
- clone 8
- concentration measureed with NanoDrop:
- clone 7 (Bba_TV_VMV_7): c = 334, 65 ng/µL
- clone 8 (Bba_TV_VMV_8): c = 183,77 ng/µL
- Plasmid need to be diluted in order to obatain the final 100pg.
- Dilution: 1:1000
- Volume needed of 1:1000 dilution of each clone:
- clone 7 (Bba_TV_VMV_7): v = 0,29 µL
- clone 8 (Bba_TV_VMV_8): v = 0,54 µL
- 100 µL aliquot of XL1-B were thawed on ice and divided into 50µL each. The proper amount of each clone was added.
Continuation: ITRs
Investigator: Hanna
In order to convert also the left ITR into RFC10 standard, the FANCY method was repeated with the newly arrived oligos (präfix and suffix).
Practical Cloning: Digestion of pAAV_MCS with AlwNI
- plasmid: P9 (270 ng/µL) = pAAV_MCS
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 (no. Lab:120) AlwNI
Comments: No.
Digestion
| components | Volume [µL] |
| DNA | 15 |
| BSA (100x) | 0.2 |
| Buffer 4 (10x) | 2 |
| Enzyme AlwNI (no.Lab:120) | 2 |
| H2O | 0.8 |
| Total volume | 20 |
- Incubation: 1 h 20 minutes
Agarose-Gel:
0.5 g Agarose, 50 mL TAE (1 %), 3 µL GELRED, at 115 Volt, running time: 50 minutes
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| P9 | 20 µl | 4 µl | 1674 bp = left ITR | 2982 bp = right ITR |
- Marker: GeneRuler ladder mix
| Marker [µL] | Sample P9 [µL] | |
|---|---|---|
| Lane | 8 | 24 |
Gelextraction
Gel measurement:
| Sample | weight | concentration |
| left ITR fragment | 0.07 g | 35.65 ng/µL |
| right ITR fragment | 0.15 g | 66.05 ng/µL |
Practical Cloning: Digestion of left ITR fragment with NotI + PstI
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 NotI-HF; Enzyme 2 PstI-HF
Comments: Due to the digestion of an gelex- and very small fragment (ITR ~ 150 bp), the whole gelex-volume was used for digestion.
Digestion
| components | Volume [µL] |
| DNA | 19 µL = 0.68 µg |
| BSA (100x) | 0.25 |
| Buffer 4 (10x) | 2.5 |
| Enzyme NotI-HF | 1 |
| Enzyme PstI-HF | 1 |
| H2O | 1.25 |
| Total volume | 25 |
- Incubation: 1 h 15 minutes
Agarose-Gel:
1.3 g Agarose, 65 mL TAE (2 %), 3 µL GELRED, at 115 Volt, running time: 45 minutes
| Sample | Sample/µl] | Loading dye (5x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| left ITR fragment | 25 µl | 4 µl | 145 bp = left ITR |
- Marker: GeneRuler ladder mix
| Marker [µL] | Sample left ITR [µL] | |
|---|---|---|
| Lane | 8 | 29 |
Gelextraction
Gel measurement:
| Sample | weight | concentration |
| left ITR | 0.09 g | 3.59 ng/µL |
Practical Cloning: Digestion of pGA14_Präfix-rITR with EcoRI + PstI
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 EcoRI-HF; Enzyme 2 PstI-HF
Comments: No.
Digestion
Plasmid: P63.1; c = 501.14 ng/µL
| components | Volume [µL] |
| DNA | 4 |
| BSA (100x) | 0.2 |
| Buffer 4 (10x) | 2 |
| Enzyme NotI-HF | 1 |
| Enzyme PstI-HF | 1 |
| H2O | 11.8 |
| Total volume | 20 |
- Incubation: 1 h 15 minutes
Agarose-Gel:
1.3 g Agarose, 65 mL TAE (2 %), 3 µL GELRED, at 115 Volt, running time: 45 minutes
| Sample | Sample/µl] | Loading dye (5x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| Präfix - right ITR | 20 µl | 3.5 µl | 165 bp = left ITR |
- Marker: GeneRuler ladder mix
| Marker [µL] | Sample Präfix-rITR [µL] | |
|---|---|---|
| Lane | 8 | 23 |
Gelextraction
Gel measurement:
| Sample | weight | concentration |
| Präfix-rITR | 0.08 g | 7.55 ng/µL |
Gel picture of left ITR fragment and Präfix-rITR fragment:
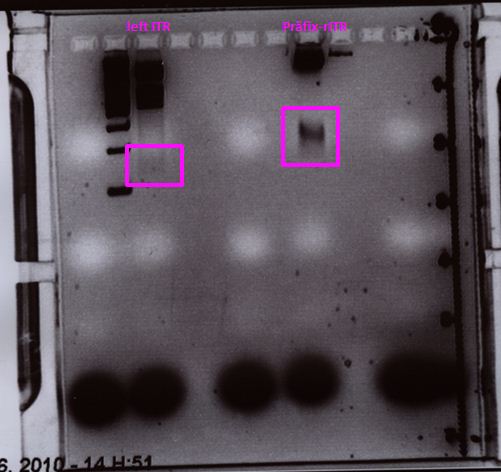
Comment: The left ITR fragment was, as expected, nearly not detectable. Nevertheless it was cut out of the gel, because the same probleme occured during the first fancy-performance, which turned out to be successful!
Practical Cloning: Digestion of pGA14 with EcoRI + NotI
- plasmid: P61 (200 ng/µL) = pGA14_Cerulean
- buffer used: 4 ; Restriction-enzymes used: Enzyme 1 = EcoRI-HF; Enzyme 2 = NotI-HF
Comments: This digested vector used used for both ligation approaches.
Digestion
| components | Volume [µL] |
| DNA | 10 |
| BSA (100x) | 0.3 |
| Buffer 4 (10x) | 3 |
| Enzyme EcoRI-HF | 1.5 |
| Enzyme NotI-HF | 1.5 |
| H2O | 13.7 |
| Total volume | 30 |
- Incubation: 1 h 15 minutes
Agarose-Gel:
0.5 g Agarose, 50 mL TAE (1 %), 3 µL GELRED, at 115 Volt, running time: 60 minutes
| Sample | Sample/µl] | Loading dye (5x)/µl | Expected size 1 (Geneious) |
|---|---|---|---|
| pGA14 | 30 µl | 4 µl | 2887 bp |
- Marker: GeneRuler ladder mix
| Marker [µL] | Sample pGA14 [µL] | |
|---|---|---|
| Lane | 8 | 34 |
Gelextraction
Gel measurement:
| Sample | weight | concentration |
| pGA14 | 0.08 g | 13.63 ng/µL |
Hybridization of ITR-Oligos
left ITR:
- 10 µL Oligo 1 (O100 = lITR_Präfix_RFC10_for bzw. O104 = rITR_Suffix_RFC10_for)
- 10 µL Oligo 2 (O101 = lITR_Präfix_RFC10_rev bzw. O105 = rITR_Suffix_RFC10_rev)
- 4 µL 100 mM TrisHCl pH8
- 8 µL 5 mM MgCl2
- 8 µL H2O
Total: 40 mL
Because some of the oligos produce strong secondary structures, we used the ORIGAMI1 program:
1) initial denaturation: 99°C, 7 minutes
2) 99°C, 1 minute
3) 73 x repetition of 2) -> -1°C, R = 0.3 °C/sec
4) hold 4°C
Ligation
1. Ligation of left ITR + Präfix + pGA14:
- Präfix-Oligos: 1 µL
- left ITR: 8 µL
- pGA14: 8 µL
- Buffer: 2 µL
- T4 Ligase: 1 µL
Total: 20 µL
2. Ligation of Präfix-rITR + Suffix+ pGA14:
- Präfix-Oligos: 1 µL
- Präfix-rITR: 8 µL
- pGA14: 8 µL
- Buffer: 2 µL
- T4 Ligase: 1 µL
Total: 20 µL
The ligation approaches were incubated at 16°C over night (thermocycler).
Biobrick of GMK
Investigators: Stefan
aim: Biobrick construction of pAAV_RFC25_gmk for further addition of TK30.
result: Transformation worked out, clones can be picked.
expected construct:
Digestion with XbaI and AgeI and ligate into vector which was digested with XbaI and AgeI as well.
PCR of gmk:
- DNA template: puB6_V5_His6_clone1 + TK/GMK; concentration: 493,7 ng/µl
- PCR program: GMK
| components | volume in µl |
| 5x Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| pmGMK_TK30_prefix_RFC25_for (082) (1:10 dilution) | 2,5 (0,5 µM) |
| pmGMK_suffix_RFC25_rev (080) (1:10 dilution) | 2,5 (0,5 µM) |
| DNA template: puB6_V5_His6_clone1 + TK/GMK (1:100) | 0,5 µL of 1:100 dilution |
| DMSO | - |
| Phusion polymerase | 0,5 |
| H2O | 33 µL |
| Total volume (e.g. 50 µl) | 50 |
Digestion of pAAV_RCF25:
- DNA-Concentration: 321,5 ng/µl
| components | volume of vector /µl |
| DNA | 4 |
| BSA (10x) | 3 |
| Buffer 4 (10x) | 3 |
| AgeI | 1 |
| XbaI | 1 |
| H2O | 18 |
| Total volume (e.g. 15,20,25,30 µl) | 30 |
Gel running of digested vector and PCR products
- 1% Agarose gel
- 3 µl Gelred, 8 µl DNA-Ladder-Mix
- 125 Volt, running time: 60 minutes
Sample of Hanna was loaded as well, see extra journal entry.
| Sample/µl | Loading dye (5x)/µl | Expected size/bp |
| Vector/ 30 | 6 | 4636 |
| gmk/ 50 | 10 | 603 |
Gelextraction
- Gel extraction performed following standard protocol provided by Qiagen
| Sample/ | Weight/g | QG buffer/µl |
| pAAV_RFC25 | 0,15 | 450 |
| gmk | 0,21 | 630 |
Digestion of PCR products
| components | volume of vector /µl |
| DNA | 30 |
| BSA (10x) | 4 |
| Buffer 4 (10x) | 4 |
| AgeI | 1 |
| XbaI | 1 |
| H2O | 0 |
| Total volume (e.g. 15,20,25,30 µl) | 40 |
Purification of PCR products, Ligation, Transformation
Purification has been followed by a different protocol (see Sven); has to be added in the standard protocols !
- Measure DNA-concentration with Nanodrop:
c(pAAV_RFC25) = 35,46 ng/µL
c(gmk) = 72,84 ng/µL
- Calculation of volume needed for ligation:
c(pAAV_RFC25) = 7,56 µL
c(gmk) = 1,44 µL
71. Labortag 27.7.2010
Cloning of linkers in pSB1C3_RFC25_CFP1.1
Investigator: Jessica
Linkers longlinker, SEG and GSAT should be cloned into the RFC25-standard
- P66: GSAT_linker_pMA 242,7 ng/µl
- P67: LonglinkerA50_pMA 230,4 ng/µl
- P71: SEG_pMA 211,8 ng/µl
- P51: pSB1C3_RFC25_CFP1.1 230,1 ng/µl
all linkers and pSB1C3_RFC25_CFP1.1 are digested with AgeI and XbaI for 2 hours
| components | volume of vector /µl (P66) | volume of insert /µl (P67) | volume of insert /µl (P71) | volume of insert /µl (P51) |
| DNA | 8,2 | 8,7 | 9,4 | 8,7 |
| BSA (10x) | 2 | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 | 2 |
| AgeI | 1 | 1 | 1 | 1 |
| XbaI | 1 | 1 | 1 | 1 |
| H2O | 5,8 | 5,3 | 4,6 | 5,3 |
| Total volume | 20 | 20 µl | 20 µl | 20 µl |
- 1% Agarose gel
- 3 µl Gelred, µl DNA-Ladder-Mix for P51, Low range for P66,67,71
- 130 Volt, running time: 45 minutes for P51, 90 Volt, running time 30 minutes for P66,67,71
| Sample/µl | Expected size/bp |
| Vector P51 | 2078 |
| Insert P66 | 126 |
| Insert P67 | 54 |
| Insert P71 | 126 |
- weight of P66 gel extract: 0,04g
- weight of P67 gel extract: 0,09g
- weight of P71 gel extract: 0,1g
- weight of P51 gel extract: 0,15g
finally called:
- pSB1C3_RFC25_longlinker P96
- pSB1C3_RFC25_GSAT P97
- pSB1C3_RFC25_SEG P98
Nanodrop
- Longlinker (P67): 2,1ng/µl
- SEG (P71): 3,7ng/µl
- GSAT (P66): 1,4ng/µl
- pSB1C3_RFC25: 29,6ng/µl
Ligation
- P51:P66 - 2,2µl:6,8µl
- P51:P67 - 2,53µl:6,47µl
- P51:P71 - 5,54µl:3,46µl
Transformation
- 4µl DNA was added to 50µl XL1b
Result sequencing of pAAV_RFC25_mGMK_TK30
Investigator: Bea
Comments: Results of the PCR and cloning experiment from lab day: 23.07.2010 (Mini-Prep). The sucessful insertion of mGMK_ZK 30 into the vector pAAV_RFC25 can be confirmed and is ready for cell culture.
Results sequencing of plasmid P81 (pAAV_RFC25_mGMK_TK30 clone 1)
Results sequencing of plasmid P82 (pAAV_RFC25_mGMK_TK30 clone 2)
Results sequencing of plasmid P83 (pAAV_RFC25_mGMK_TK30 clone 3)
Quickchange @ Pst in RepX 68 and RepX 78
Investigator: Kira
PCR program: PstI
| components | volume in µl |
| 10x Phusion HF buffer | 2.5 µl |
| 10 mM dNTP mix | 0.5 µl |
| __310 ____primer_for (1:10 dilution) | 0,39 µl |
| __310____primer_rev (1:10 dilution) | 0,39 µl |
| DNA template (1:20 dilution) | 0,5 µl |
| DMSO | 0 µl |
| Phusion polymerase | 0.5 µl |
| H2O | 20,22 µl |
| Total volume (e.g. 50 µl) | 25 µl |
| cycles | temperature | time |
| 1 | 95 C | 2 min |
| 20 | 95 C | 30 sec |
| 66 C | 1 min | |
| 68 C | 5 min |
- The PCR-products were digested with 0.5 µl DpnI
- Transformation was performed according to the standard protocol in XL1B cells
Site-directed mutagenesis of pAAV_RC_PstI_310+4073 (pAAV_RC_1.1) with BamHI and SalI
Investigator: Adrian, Bea
Comments: Site-directed mutagenesis was perfomed with pAAV_RC_PstI_310+4073 (pAAV_RC_1.1) in order to delete BamHI and SalI in the plasmid. These restriciton sites need to be removed because they are the single-cutter in the loops which will be used for targeting.
Protocol: Different PCR programs were used for performing the Quickchange site-directed mutagenesis protocol.
- Longer elongation/extension time
- Shorter elongation/extension time
1. PCR reaction with PCR program with longer elongation time:
DNA template used: pAAV_RC_PstI_310+4073 (pAAV_RC_1.1) = P87
A dilution of 1:20 was prepared and it was calculated the amount of DNA template needed for each site-directed mutagenesis reaction.
| components | volume in µl | volume in µl| |
| 10x Phu Ultra HF II buffer | 2.5 µl | 2.5 µl |
| 10 mM dNTP mix | 0.5 µl | 0.5 µl |
| primer_for (1:10 dilution) | O68= SDM SalI: 0,67 µl | O66= SDM BamHI: 0,68 µl |
| primer_rev (1:10 dilution) | O69= SDM SalI: 0,67 µl | O67= SDM BamHI: 0,72 µl |
| P87 (DNA template) (1:20 dilution) | 0,83 µl | 0,83 µl |
| DMSO | - | - |
| Phusion polymerase | 0.5 µl | 0.5 µl |
| H2O | 19,33 µl | 18,77 µl |
| Total volume (e.g. 50 µl) | 25 µl | 25 µl |
Used PCR programs for both SDMs:
Comment: Program has a long extension time because Quikchange Kit (Stratagene) recommends 1 minute per kb. Our vector has a length of 7300 bp therefore 7 minutes and 30 seconds were chosen for proper elongation of the whole vector.
| Rounds | Temperature | Time |
| 1 | 95°C | 2' |
| 20 | 95°C | 30 |
| 60°C | 1' | |
| 68°C | 7 min 30 sec | |
| Hold | 4°C |
2. PCR reaction with PCR program with shorter elongation time:
DNA template used: pAAV_RC_PstI_310+4073 (pAAV_RC_1.1) = P87
A dilution of 1:20 was prepared and it was calculated the amount of DNA template needed for each site-directed mutagenesis reaction.
| components | volume in µl | volume in µl| |
| 10x Phu Ultra HF II buffer | 2.5 µl | 2.5 µl |
| 10 mM dNTP mix | 0.5 µl | 0.5 µl |
| primer_for (1:10 dilution) | O68= SDM SalI: 0,67 µl | O66= SDM BamHI: 0,68 µl |
| primer_rev (1:10 dilution) | O69= SDM SalI: 0,67 µl | O67= SDM BamHI: 0,72 µl |
| P87 (DNA template) (1:20 dilution) | 0,83 µl | 0,83 µl |
| DMSO | - | - |
| Phusion polymerase | 0.5 µl | 0.5 µl |
| H2O | 19,33 µl | 18,77 µl |
| Total volume (e.g. 50 µl) | 25 µl | 25 µl |
Used PCR programs for both SDMs:
Comment: Quikchange Protocol (Stratagene) recommends 30 seconds per kb chosing a elongation temperature of 68°C. Our vector has a length of 7300 bp therefore 4 minutes were chosen for proper elongation of the whole vector. In order to see wheter this PCR program works aswell the same PCR ingredients were chosen as it can be seen in the table above.
| Rounds | Temperature | Time |
| 1 | 95°C | 2' |
| 20 | 95°C | 30 |
| 60°C | 1' | |
| 68°C | 4' | |
| Hold | 4°C |
Summary: Four site-directed mutagenesis have been performed with the plasmid pAAV_RC_PstI_310+4073 (pAAV_RC_1.1) = P87.
- Site-directed mutagenesis in order to delete restriction site BamHI
- with shorter elongation time (modified protocol)
- with longer elongation time
- Site-directed mutagenesis in order to delete restriction site SalI
- with shorter elongation time (modified protocol)
- with longer elongation time
- The samples were digested with 0,5µL DpnI after site-directed mutagenesis were performed in order to remove bacterial DNA template.
- Trafos have been performed with all four samples. Trafo plates are containing ampicillin.
Cell culture
Investigator: Adrian
- 3 x 6er dishes for Transduction were seeded and passaged(P14)
ITRs: Trafo of overnight-ligation approaches
Investigator: Hanna
Comments: Cloning approach was continued: RFC10-Präfix_rightITR was ligated to RFC10-Suffix and pGA14 and leftITR was ligated to RFC10-Präfix and pGA14 over night. Trafo of these samples was performed in order to pick clones tomorrow.
Trafo was performed following the standard protocol. Abberance: 4 µL DNA was added to BL21 cells.
PCR: beta-Globin
Investigator: Hanna
Because sequencing delivered, that cloning of the beta-globin intron into pSB1C3 didn't work (Cerulean was still in the vector), the PCR was performed one more time:
| components | volume [µl] |
| 5x Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| beta-globin_primer_for | 2,5 |
| beta-globin_primer_rev | 2,5 |
| DNA template | 1 ng = 2.8 µL of 1:1000 dilution of pAAV_MCS, c=360 ng/µL |
| DMSO | - |
| Phusion polymerase | 0,5 |
| H2O | 30.7 |
| Total volume (e.g. 50 µl) | 50 |
PCR program:
- 98°C, 1'
- 98°C, 30
- 63°C, 25
- 72°C, 15
-> 8x
- 98°C, 30
- 72°C, 20
-> 20x
- 1 x 72°C, 5'
- Hold: 4°C
Repetition of PCR for hGH BioBrick Production
Investigator: Bea
Comments: Sequencing of pSB1C3_hGH did not reveal any insertion of hGH into pSB1C3. Instead of hGH CFP was found in the vector indicating that undigested or partially digested vector religated. Therefore a new pCR for generating the BioBrick was performed over night.
PCR reaction:
| components | volume [µl] |
| 5x Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| hGH_primer_for | 2,5 |
| hGH-globin_primer_rev | 2,5 |
| DNA template | 1 ng = 2.8 µL of 1:1000 dilution of pAAV_MCS, c=360 ng/µL |
| DMSO | - |
| Phusion polymerase | 1 |
| H2O | 30.7 |
| Total volume (e.g. 50 µl) | 50 |
Used PCR programs:
Comment: PCR will be performed over night.
| Rounds | Temperature | Time |
| 1 | 98°C | 1' |
| 8 | 98°C | 30 |
| 58°C | 25 | |
| 72°C | 12 | |
| 17 | 98°C | 30 |
| 69°C | 25 | |
| 72°C | 12 | |
| Hold | 4°C |
Picking clones from pAAV_RFC25_mGMK
Investigator: Bea
Comment: Trafo plate looked good. On control plate only few clones grown in contrary to the XL1-B{pAAV_RFC25-mGMK} plate. Two clones were picked.
- 10 mL containing 10µL ampiciliin were inoculated.
- Put into 37°C room on shaker.
Picking clones from Retrafo of pTV_CMV clone 7 and clone 8
Investigator: Bea
Comment: Trafo plates looked good. Enough clones grown. Two clones of each plate were picked
- 10 mL containing 10µL ampiciliin were inoculated with each clone respectively.
- Put into 37°C room on shaker.
72. Labortag 28.7.2010
Results Trafo pAAV_RC_1.1_BamHI and pAAV_RC_1.1_SalI
Investigator: Bea
Results: Too many bacteria grown resulting in a bacterial lawn. Possible reasons could be some problems with the new prepared ampicillin because trafo from Hanna containing the ITRs had the same results. Trouble shooting done by Hanna.
Proceed with BioBrick Production of hGH and beta-globin
Investigator: Bea
Comments: PCR was started yesterday and conducted over night.
Proceeding:
- Prepare 1% agarose gel and load samples on gel.
- Run time: 45 minutes
Result of gel:
Marker did not resolute very good. Expected sizes of PCR products:
- hGH = 510 bp
- beta globin intron: 524 bp
It can be seen (or estimated) that two intensive bands are visible at around 500 bp. Therefore these two bands were cut out and a gel extraction was performed. The lower visible band at around 100 bp of hGH is an additional band generated at the pCR reaction. The two intensive blurred bands belong to the loading dye added to the samples.
After gel extraction was performed, the PCR prdoucts hGH and beta-globin were digested:
DIGESTION of PCR products
| components | hGH/µl | beta-globin /µL |
| pSB1C3_CFP | 29 | 29 |
| BSA (100x) | 0,4 | 0,4 |
| Buffer 4 (10x) | 4 | 4 |
| XbaI | 1 | 1 |
| PstI | 1 | 1 |
| H2O | 4,6 | 4,6 |
| Total volume (e.g. 15,20,25,30 µl) | 40 | 40 |
- Incubate for 2h at 37°C
- Perform PCR purification following standard protocols
- Elute in 20µL EB-buffer, put on 50°C for 3 minutes before centrifugating
- Measure DNA with NanoDrop:
- c(hGH) = 58,14 ng/µL
- c(ß-globin) = 83,10ng/µL
In the meantime prepare the digestion of vector pSB1C3_CFP (P51)
DIGESTION of pSB1C3_CFP
| components | volume /µl |
| pSB1C3_CFP | 5 |
| BSA (10x) | 2 |
| Buffer 4 (10x) | 2 |
| XbaI | 1 |
| PstI | 1 |
| H2O | 9 |
| Total volume (e.g. 15,20,25,30 µl) | 20 |
- Incubate for 2h at 37°C
- Load sample on 1% preparative agarose gel
- Run for 45 minutes at 110 V
Result of gel:
The vector was digested with XbaI and PstI. Expected sizes have been:
- pSB1C3 = 2072bp
- CFP: 732bp
It can be seen that the vector pSB1C3_RFC25_CFP was digested and two visible bands can be seen on the gel. The lower band corresponds to the CFP fragment (~700 - 800 bp), the higher band corresponds to the plasmid backbone at around 2000-3000 bp. The higher band was cut out and extracted by gel extraction.
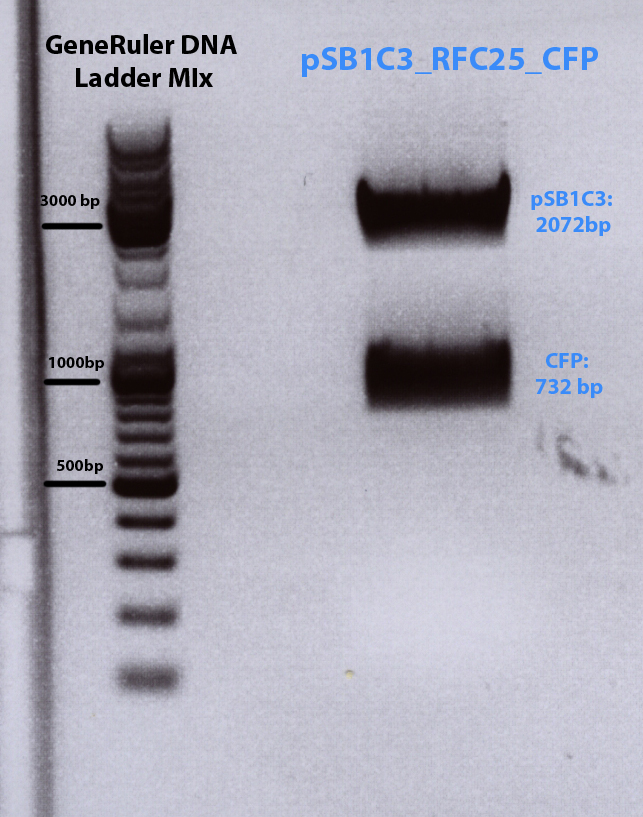
- Perform gel extraction and measure DNA with NanoDrop
- c(psb1C3) = 29,52 ng/µL
LIGATION of pSB1C3 with hGH and beta-globin
Calculated ratio of pSB1C3 (vector) and hgh(insert) or betaglobin (insert)

The ligated samples
- pSB1C3_hGH
- pSB1C3_betaglobin
were transformed with XL1B and incubated on agar-plates containing chloramphenicol over night at 37°C.
Results of ITR Trafo
Investigator: Hanna
Because (as mentioned above) there was a bacteria lawn on the right-ITR plate and many colonies on the left-ITR plate three different approaches were performed:
1. Trafo of rightITR-RFC25_pGA14 and leftITR-Präfix_pGA14 was conducted one more time:
Abberances:
- 3µL DNA-sample was added to 50 µL cells (BL21).
- Cells were incubated on ice for just 10 minutes.
- Heat shock: 42 seconds at 42°C.
- Incubation for just 35 minutes at 37°C, rotating (800 rpm).
- Agar plates were prepared with Lab-Ampicillin.
Incubation at 37°C -> Picking clones in the evening.
2. Control: Bacteria were taken from the right-ITR-lawn an resuspended in 100 µL DYT. They were plated on "LabAmp-plates" and stored at 37°C.
-> If something is wrong with our Ampicillin the non-transformed bacteria have progeny-advantages and will crowd out the transformed cells. This means: If there will be no colonies detectable on the new plates, something is wrong with our Ampicillin!
Analysis: Tonight.
3. Picking clones of the left ITR plate: Two large and two small clones were picked from the left ITR plate and each was resuspened in 50 µL DYT. An "LabAmp-plate" was quartered and the four resuspened colonies were individualized each on one quarte of the plate.
Analysis: Tonight.
Trafo evaluation Rep 68&78 Pst
Investigator: Kira
Both plates Rep 68 & Rep 78 contain lots of colonies. Approx. 4 colonies from each plate will be picked and inoculated for Mini prep tomorrow.
Cellculture
Investigator: Jessica
HEK were split according to the standard protocol in 5 flasks
Mini-Prep and Test digestion of pAAV_RFC25_mgmk and pTV_CMV_7 and pTV_CMV_8
Investigator: Stefan
Comments:
mgmk was brought into iGEM standard vector. To verify the construct it will be sent for sequencing.
pTV_CMV_7 and pTV_CMV_8 are clones we recieved from the iGEM 2008 team. Because there are mistakes in the prefix and suffix a PCR will have to be done. They will be sent for sequencing as well.
Mini-Prep of pAAV_RFC25_mgmk clone 1 and 2: After performing Mini-Preps concentration was measured with Nanodrop:
- pAAV_RFC25_mgmk clone 1: c = 326,0 ng/µl
- pAAV_RFC25_mgmk clone 2: c = 411,3 ng/µl
Mini-Prep of pTV_CMV_7 clone 1 and 2: After performing Mini-Preps concentration was measured with Nanodrop:
- pTV_CMV_7 clone 1: c = 378,2 ng/µl
- pTV_CMV_7 clone 2: c = 412,9 ng/µl
Mini-Prep of pTV_CMV_8 clone 1 and 2: After performing Mini-Preps concentration was measured with Nanodrop:
- pTV_CMV_8 clone 1: c = 337,6 ng/µl
- pTV_CMV_8 clone 2: c = 366,6 ng/µl
Test digestion:
pAAV_RFC25_mgmk was digested with XbaI and AgeI.
pTV_CMV_7 and pTV_CMV_8 were digested with XbaI and PstI.
- P99 = pAAV_RFC25_mgmk clone 1
- P100 = pAAV_RFC25_mgmk clone 2
- P101 = pTV_CMV_7 clone 1
- P102 = pTV_CMV_7 clone 2
- P103 = pTV_CMV_8 clone 1
- P104 = pTV_CMV_8 clone 2
| components | volume of P99/µl | volume of P100/µL | volume of P101/µL | volume of P102/µl | volume of P103/µL | volume of P104/µL |
| DNA | 2,5 | 2 | 2,5 | 2 | 2,5 | 2,5 |
| BSA (10x) | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 |
| Buffer 4 (10x) | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 |
| XbaI | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 |
| AgeI | 0,5 | 0,5 | / | 0/ | / | / |
| PstI | / | / | 0,5 | 0,5 | 0,5 | 0,5 |
| H2O | 8,5 | 9 | 9 | 8,5 | 8,5 | 8,5 |
| Total volume /µl | 15 | 15 | 15 | 15 | 15 | 15 |
Expected sizes:
- mgmk: 603bp
- pAAV_RFC25: 4636bp
- CMV: 679bp
- pTV: 3408bp
Results of samples loaded 1% agarose gel and ran ~50 minutes at 115 V:
Sequencing
70ng/µL were sent to GATC from each clone. They were labeled:
- SB 1 (pAAV_RFC25_mgmk clone 2 (P100))
- SB 2 (pTV_CMV_7 clone 1 (P101))
- SB 3 (pTV_CMV_8 clone 1 (P103))
For SB 1 there was sent the GOI_forward primer (O30) as well.
Site-directed mutagenesis of SspI and PvuII in pSB1C3_CFP
Investigator: Jessica
Comments: Site-directed mutagenesis was perfomed with pSB1C3_CFP in order to delete SspI and PvuII in the plasmid. These restriciton sites need to be removed because they are also in the loop we want to modify.
PCR reaction with PCR program with SspI and PvuII:
DNA template used: ppSB1C3_CFP = P51
| components | volume in µl | volume in µl| |
| 10x PfuUltra II buffer | 2.5 µl | 2.5 µl |
| 10 mM dNTP mix | 0.5 µl | 0.5 µl |
| primer_for (1:10 dilution) | O107= SDM SspI: 0,55 µl | O109= SDM PvuII: 0,58 µl |
| primer_rev (1:10 dilution) | O106= SDM SspI: 0,55 µl | O108= SDM PvuII: 0,59 µl |
| P51 (DNA template) ) | 0,43 µl | 0,43 µl |
| DMSO | - | 0,5µl |
| PfuUltra II Fusion | 0.5 µl | 0.5 µl |
| H2O | 16,1 µl | 15,53 µl |
| Total volume (e.g. 50 µl) | 25 µl | 25 µl |
Used PCR programs for SDMs:
1) PvuII
| Rounds | Temperature | Time |
| 1 | 95°C | 2' |
| 20 | 95°C | 30 |
| 78°C | 1' | |
| 68°C | 2 min 50 sec | |
| Hold | 4°C |
2) SspI
| Rounds | Temperature | Time |
| 1 | 95°C | 2' |
| 20 | 95°C | 30 |
| 79°C | 1' | |
| 68°C | 2 min 40 sec | |
| Hold | 4°C |
- Transformation was prepared according to the standard protocol
Affibody
Investigator: Hanna
In order to target the EGF-receptor (EGFR/ErbB-1/Her1) an affibody molecule can be fused to the N-terminus of e.g. VP2, which is expressed in trans. The ZEGFR:1907 was chosen for this approach. Because just the amino acid sequence is published by Friedman et al., 2008, the sequence was translated with Mr. Gene's help :) and optimized for mammalian expression.
Strategy:
Picking clones from Trafo pSB1C3_RFC25_longlinker, pSB1C3_RFC25_GSAT, pSB1C3_RFC25_SEG
Investigator: Jessica
Comment: Trafo plates looked good. Enough clones grown. Two clones of each plate were picked
- 10 mL containing 10µL chloramphenicol were inoculated with each clone respectively.
- Put into 37°C room on shaker.
Preparation for competent XL1b
Investigator: Jessica 15 ml DYT was prepared with tetracyclin and XL1b from glycerolstock and incubates over night. goes on on thursday.
ReTrafo of pGL3_hTERT
Investigator: Bea
Comment: pGL3_hTERT received from Kira. Contains ampiciliin restitance. Retrafo was performed in order to obtain enough DNA to conduct PCR for add-on tails.
Re-transformation:
- Construct used: pGL3_hTert
- concentration c = 276ng/µL
- Plasmid need to be diluted in order to obatain the final 100pg.
- Dilution: 1:1000
- Volume needed of 1:1000 dilution of pGL3_hTert:
- v = 0,37µL
- 100 µL aliquot of XL1-B were thawed on ice and the plasmid DNA was added.
Trafo of PAAV_RC_1.1 SDM of BamHI and SalI
Investigator: Bea
Comment: Trafo did not work (see above) therefore th etRafo have been performed again
Picking clones from the 2. Trafo of left and right ITR (Hanna)
Investigator: Jessica
Comment: Trafo plates looked good. Enough clones grown. Four clones of each plate were picked
- 10 mL containing 10µL Amp were inoculated with each clone respectively.
- Put into 37°C room on shaker.
Comment @Hanna: also on control plate are clones grown... not on the quartered plate. back in the 37°C room
73. Labortag 29.7.2010
Mini-Prep and Test digestion of Rep 68 and Rep 78
Investigator: Kira
3 clones from each plate were picked and inoculated. The mini prep DNA extraction revealed followed DNA concentrations:
Rep 68 clone 1: 470 ng/μl clone 2: 556,15 ng/μl clone 3: 686,40 ng/μl
Rep 78 clone 1: 566,78 ng/μl clone 2: 519,68 ng/μl clone 3: 521,88 ng/μl
All 6 samples were test digested with PstI to check if the site directed mutagenesis was successful.
| Components | Volume/µL | Mastermix |
| DNA | Variable (800 ng) | - |
| BSA (10x) | 1 | 6 |
| Buffer no. 4 (10x) | 1 | 6 |
| Enzyme 1 ( PstI ) | 0,5 | 3 |
| H2O | variable | - |
| Total volume | 10 | - |
- Incubation: 1h @ 37 C
1% agarose gel
- Marker: GeneRuler ladder mix
| Marker /µl | Sample Rep 68 clone 1 /µl | Sample Rep 68 clone 2 /µl | Sample Rep 68 clone 3 /µl | Sample Rep 78 clone 1 /µl | Sample Rep 78 clone 2 /µl | Sample Rep 78 clone 3 /µl | |
|---|---|---|---|---|---|---|---|
| Lane | 7 | 12 | 12 | 12 | 12 | 12 | 12 |
Results sequencing of pTV_CM clone 7 and 8 (received from iGEM team 2008)
Investigator: Bea
Comment: Sequencing results obtained from the Mini-Prep performed yesterday from the constructs pTV_CMV_7 and pTV_CMV_8 received of the iGEM Freiburg team 2008. A Retrafo has been performed and obtained plasmids of this retrafo have been send for sequencing.
Results: Seqeuncing reads look good. Plasmids contain expected CMV promoter which is already in the iGEM RFC10 standard but with one missing "G" in the prefix.
Results prefix RFC10:
Results pTV_CMV_7:
Results pTV_CMV_8:
Sequencing results of pAAV_RFC25_mGMK
Investigator: Bea
Comment: Sequencing results of pAAV_RFC25_mGMK. A PCR has been perfomed in order to obtain the desired mGMK construct in the RFC25 standard. After Mini-Prep has been perfomed clone 1 has been sent to GATC for sequencing.
Conclusion: The PCR amplification with designed primers to insert prefix and suffix in RFC25 standard can be verified. No nucleotide transitions can be detected. The prefix and suffix are in the designed standards. Additionally it can be confirmed that the mGMK was inserted successfully into the pAAV_RFC25 plasmid and do NOT contain any iGEM restriction sites When receiving the ordered tk30 from Mr.Gene it can be fused to the mGMK and will the be ready for using and sending to the IGEM registry (maybe this needed to be cloned into pSB1C3).
Stock solution of Ampicilin
Investigators: Johannes, Stefan
0,3g Ampicilin were dissolved in 3ml Millipore H20. PCR tubes were filled with 60 µl each and stored at -20 °C.
Mini-Prep and Test digestion of ITR constructs
Investigator: Hanna
The RFC10-Prefix was fused to the left ITR and cloned into the pGA14_Cerulean plasmid. 4 clones of the second trafo (the first trafo didn't work due to wrong preparation of the Ampicillin stock solution, see 28.7.2010) were picked yesterday. Mini-Prep was performed today.
Plasmid-no.: (pGA14_10Prefix_lITR)
P111: 321.91 ng/µL
P112: 489.96 ng/µL
P113: 341.49 ng/µL
P114: 402.3 ng/µL
Glycerol stock were prepared: B85.1 - B85.1.
The "Intermediate-Step-Suffix" was fused to the RFC10-Prefix-rITR construct and cloned into pGA14. 4 clones of the second trafo were picked yesterday. Mini-Prep was performed today.
Plasmid-no.: (pGA14_10Prefix_PreSuffix_rITR)
P115: 401.07 ng/µL
P116: 291.7 ng/µL
P117: 312.98 ng/µL
P118: 326.78 ng/µL
Glycerol stock were prepared: B84.1 - B84.1.
Test digestion
- buffer used: 4 ; Restriction-enzymes used: Enzyme NotI-HF
- Plasmid: P111 - P118
Pipetting scheme for for each digestion:
| Components | Volume/µL |
| DNA | 4 µL |
| BSA (100x) | 0.1 |
| Buffer no.4 (10x) | 1 |
| Enzyme NotI-HF | 1 |
| H2O | 3.9 |
| Total volume | 10 |
- Incubation: 55 minutes
Agarose-Gel:
0.8 g Agarose, 55 mL TAE (1.45 %), 3 µL GELRED, at 115 Volt, running time: 35 minutes
| Sample | Sample/µl] | Loading dye (5x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| pGA14_Prefix_lITR clone 1 | 10 µl | 2 µl | ~ 150 bp | ~ 2900 bp |
| pGA14_Prefix_lITR clone 2 | 10 µl | 2 µl | ~ 150 bp | ~ 2900 bp |
| pGA14_Prefix_lITR clone 3 | 10 µl | 2 µl | ~ 150 bp | ~ 2900 bp |
| pGA14_Prefix_lITR clone 4 | 10 µl | 2 µl | ~ 150 bp | ~ 2900 bp |
| pGA14_Prefix_PreSuffix_rITR clone 1 | 10 µl | 2 µl | 145 bp | ~ 2900 bp |
| pGA14_Prefix_PreSuffix_rITR clone 2 | 10 µl | 2 µl | 145 bp | ~ 2900 bp |
| pGA14_Prefix_PreSuffix_rITR clone 3 | 10 µl | 2 µl | 145 bp | ~ 2900 bp |
| pGA14_Prefix_PreSuffix_rITR clone 4 | 10 µl | 2 µl | 145 bp | ~ 2900 bp |
- Marker: GeneRuler ladder mix
| Marker /µl | Sample pGA14_Prefix_lITR clone 1 /µl | Sample pGA14_Prefix_lITR clone 2 /µl | Sample pGA14_Prefix_lITR clone 3 /µl | Sample pGA14_Prefix_lITR clone 4 /µl | Sample pGA14_Prefix_PreSuffix_rITR clone 1 /µl | Sample pGA14_Prefix_PreSuffix_rITR clone 2 /µl | Sample pGA14_Prefix_PreSuffix_rITR clone 3 /µl | Sample pGA14_Prefix_PreSuffix_rITR clone 4 /µl | |
|---|---|---|---|---|---|---|---|---|---|
| Lane | 6 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 |
Comments:Because samples can be sent to GATC just until 7 p.m., gel run was checked already after 15 minutes:
No bands can be detected at the expected sizes. The fragment size seems to be inbetween the expectation of either 150 and 750 bp. Further on there is also an additional band visible. Because of that it's not possible to say, whether cloning worked. Therefore the three clones marked in the picture were sent to GATC for sequencing:
- pGA14_Prefix_lITR clone 1: 6.5 µL plasmid + 23.5 µL H2O = "Hanna 1"
- pGA14_Prefix_lITR clone 4: 5.2 µL plasmid + 24.8 µL H2O = "Hanna 2"
- pGA14_Prefix_PreSuffix_rITR clone 1: 5.2 µL plasmid + 24.8 µL H2O = "Hanna 3"
Picking clones of pGL3_hTERT
Investigator: Bea
Comments: Clones will be picked from trafo plate in order to obatin single colonies in order to perform Mini-prep tomorrow.
- Bacterial strain used: XL1-B
- Two clones were picked of trafo plate
- Inoculating of 10 mL DYT medium containing 10µL ampicillin
Picking clones of pAAV_RC_1.1 SDM of BamHI and SalI
Investigator: Bea
Comments: All trafo plates looked good, enough clones grew, aside from trafo plate XL1-B {pAAV_RC_1.1_SalI}: long PCR, where no clones grew on plate. Clones will be picked from each trafo plate in order to obatin single colonies in order to perform Mini-Prep tomorrow.
- Bacterial strain used: XL1-B
- Two clones were picked of each trafo plate
- XL1-B {pAAV_RC_1.1_BamHI}: long PCR
- XL1-B {pAAV_RC_1.1_BamHI}: short PCR
- XL1-B {pAAV_RC_1.1_SalI}: short PCR
- Inoculating of 10 mL DYT medium containing 10µL ampicillin
Picking clones of pSB1C3_hGH and pSB1C3_betaglobin
Investigator: Bea
Comments: Clones will be picked from trafo plate in order to obatin single colonies in order to perform Mini-Prep tomorrow.
- Bacterial strain used: XL1-B
- Two clones were picked of each trafo plate
- XL1-B {pSB1C3_hGH}
- XL1-B {pSB1C3_betaglobin}
- Inoculating of 10 mL DYT medium containing 10µL chloramphenicol
PCR with pTV_CMV with designed oligos for amplifying CMV in RFC10 standard
Investigator: Bea
Comments: Retrafo of th epTV_CMv constructs has been performed and seqeuncing revealed that the plasmid contains the CMV promoter. Therefore, PCR in order to add prefix and suffix in RFC10 standard can be performed.
- Plasmid used: pTV_CMV_7 clone 2 (P102)
- Plasmid has to be diluted to provide the possibilty of pipetting the volume.
- 1:1000 dilution of 412ng/µL --> c(new) = 0,412ng/µL
- Primer used:
- pCMV_prefix_RFC10_for (1:10)
- pCMV_suffix_RFC10_rev (1:10)
PCR reaction:
| components | volume /µl |
| 5x Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| pCMV_prefix_for_RFC10 (1:10) | 2,5 |
| pCMV_suffix_for_RFC10 (1:10) | 2,5 |
| pTV_CMV_7 clone 1 (P102) | 1.8 ng = 4,5 µL of 1:1000 dilution of P102, c=412 ng/µL |
| DMSO | - |
| Phusion polymerase | 0,5 |
| H2O | 29 |
| Total volume (e.g. 50 µl) | 50 |
Used PCR programs:
Comment: PCR will be performed over night.
| Rounds | Temperature | Time |
| 1 | 98°C | 1' |
| 30 | 98°C | 15 |
| 70°C | 25 | |
| 72°C | 12 | |
| Hold | 4°C |
Mini-Prep and Test digestion of pSB1C3_RFC25_longlinker, pSB1C3_RFC25_SEG and pSB1C3_RFC25_GSAT
Investigator: Jessica
Nanodrop
- pSB1C3_RFC25_longlinker clone1: 187,8 ng/µl P105
- pSB1C3_RFC25_longlinker clone2: 197,4 ng/µl P106
- pSB1C3_RFC25_SEG clone1: 185,6 ng/µl P107
- pSB1C3_RFC25_SEG clone2: 182,6 ng/µl P108
- pSB1C3_RFC25_GSAT clone1: 179,5 ng/µl P109
- pSB1C3_RFC25_GSAT clone2: 202,5 ng/µl P110
Test digestion 1µg DNA
| components | P105 /µl | P107 /µl | P109 /µl |
| DNA | 5,3 | 5,3 | 5,6 |
| BSA (10x) | 1 | 1 | 1 |
| Buffer 4 (10x) | 1 | 1 | 1 |
| H2O | 2,2 | 2,2 | 1,9 |
| Total volume (e.g. 15,20,25,30 µl) | 10 | 10 | 10 |
agarose gel: 2%, digestion: 45 minutes, 37°C
P105, P107 and P109 are sent for sequenzing with Reverse Primer (VR2)
Comments:this is no satisfying result. we can't imagine what this bands stand for. maybe the CFP is still in the pSB1C3 and the digestion of the pSB1C3_RFC25_CFP didn't work. P105, P107 and P109 are sent for sequenzing to look for the CFP respectively the linkers
Picking clones of pSB1C3_CFP w/o SspI and PvuII
Investigator: Jessica
- Bacterial strain used: XL1-B
- Four clones were picked of trafo plate
- Inoculating of 10 mL DYT medium containing 10µL chloramphenicol
Hanna will do the Mini Prep tomorrow
Inoculation of pSB1C3_RFC25_CFP
Investigator: Jessica
- 200ml DYT was prepared with 200µl chloramphenicol and the glycerol stock no. B39
- stored in the 37°C room over night for Midi Prep tomorrow
74. Labortag 30.7.2010
Continuation of PCR with pTV_CMV and digestion of pSB1C3
Investigator: Bea
Comments: Retrafo of the pTV_CMv constructs has been performed and sequencing revealed that the plasmid contains the CMV promoter. Therefore, PCR was be performed overnight. After PCR the PCR product was loaded on a 1% preparative agarose gel. While PCR was separated by the agarose gel, digestion of pSB1C3_CFP was performed.
Proceeding:
- Prepare 1% agarose gel and load samples on gel.
- Run time: 45 minutes
Result of gel:
Expected sizes of PCR products:
- CMV = 681 bp
After gel extraction was performed, the PCR prdouct CMV was digested:
DIGESTION of PCR products
| Components | Volume/µL |
|---|---|
| PCR product: CMV | 29 |
| BSA (100x) | 0,4 |
| Buffer 4 (10x) | 4 |
| Enzyme XbaI | 1 |
| Enzyme PstI | 1 |
| H20 | 4,6 |
| Total volume | 40 |
- Incubate for 2h at 37°C
- Perform PCR purification following standard protocols
- Elute in 20µL EB-buffer, put on 50°C for 3 minutes before centrifugating
- Measure DNA with NanoDrop:
- c(CMV) = 87,4ng/µL
In the meantime prepare the digestion of vector pSB1C3_CFP (P51)
DIGESTION of pSB1C3_CFP
| Components | Volume/µL |
|---|---|
| pSB1C3_CFP | 6 |
| BSA (10x) | 2 |
| Buffer 4 (10x) | 2 |
| Enzyme XbaI | 1 |
| Enzyme PstI | 1 |
| H20 | 8 |
| Total volume | 20 |
- Incubate for 2h at 37°C
- Load sample on 1% preparative agarose gel
- Run for 45 minutes at 110 V
Result of gel:
The vector was digested with XbaI and PstI. Expected sizes have been:
- pSB1C3 = 2072bp
- CFP: 732bp
- Proceed with ligation of pSB1C3 and CMV
- v (vector) = 7,37µL
- v (insert)= 1,63µL
- Trafo has been performed with XL1B cells.
- 100µL if XL1B cells have been transformed with 2µL of pSB1C3_CMV
- Transformed cells have been plated on agar plates containing chloramphenicol
Next steps: Picking clones of trafo plate and performing Mini-Prep. When all BioBricks are ready conduct assembly strategy and test constructs.
Repetition of test digestion of Rep 68 and Rep 78
Investigator: Kira
| Components | Volume/µL | Mastermix |
| DNA | Variable (800 ng) | - |
| BSA (10x) | 1 | 6 |
| Buffer no. 4 (10x) | 1 | 6 |
| Enzyme 1 ( PstI ) | 0,5 | 3 |
| H2O | variable | - |
| Total volume | 10 | - |
- Incubation: 1h @ 37 C
1% agarose gel
- Marker: GeneRuler ladder mix
1/3 of the digested solution was diluted in 2/3 water because the agarose gel from yesterday showed very distinct bands
| Marker /µl | Sample Rep 68 wt | Sample Rep 68 clone 1 /µl | Sample Rep 68 clone 2 /µl | Sample Rep 68 clone 3 /µl | Sample Rep 78 clone 1 /µl | Sample Rep 78 clone 2 /µl | Sample Rep 78 clone 3 /µl | Sample Rep 78 wt | |
|---|---|---|---|---|---|---|---|---|---|
| Lane | 7 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 |
TAE buffer was changed before start, the samples have been diluted to 1/3 but the bands are still very distinct. However, it is possible to analyse the gel. Rep 68 contains no PstI restriction sites anymore, while Rep 78 shows one band, which might be due to the additional PstI restiction site.
clone1_Rep68 and clone1_ Rep78 were sent for sequencing
Continuation: left ITR
Investigator: Hanna
GATC didn't manage to do the sequencing of the left ITR-Präfix construct until now. This can be interpreted as positive news, because this increases the fact that the ITR is in the plasmid (very strong secondary structures!). Therefore cloning will be continued today:
Practical Cloning:
Digestion of pGA14_Cerulean (P61) and of pGA14_10Präfix-lITR (P111)
| components | volume of pGA14_Cerulean (P61) /µl | volume of pGA14_10Präfix-lITR (P111) /µl |
| DNA | 7.5 | 9.3 |
| BSA (100x) | 0.2 | 0.2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme 1 | PstI-HF: 1 | NotI-HF: 1 |
| Enzyme 2 | XbaI: 1 | XbaI: 1 |
| H2O | 8.3 | 6.5 |
| Total volume | 20 | 20 |
- Incubation: ~2 h
Agarose-Gel:
P61:
0.5 g Agarose, 50 mL TBE!!! (1 %), 3 µL GELRED, at 115 Volt, running time: 45 minutes
P111:
0.8 g Agarose, 50 mL TBE!!! (1.45 %), 3 µL GELRED, at 115 Volt, running time: 35 minutes
| Sample | Sample/µl] | Loading dye (5x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| P61 | 20 µl | 5 µl | 2896 bp | ~ 750 bp |
| P111 | 20 µl | 5 µl | 151 bp | ~ 2890 bp |
- Marker: GeneRuler ladder mix
Gelextraction
Gel measurement:
| Sample | Concentration |
| P61 | 12.44 ng/µL |
| P111 | 11.21 ng/µL |
Oligo-Hybridization
RFC10-Suffix of left ITR:
- 10 µL Oligo 1 (O102)
- 10 µL Oligo 2 (O103)
- 4 µL 100 mM TrisHCl pH8
- 8 µL 5 mM MgCl2
- 8 µL H2O
Total: 40 mL
Thermocycler program: ORIGAMI1
1. 99°C, 7'
2. 99°C, 1'
73 x, -1°C; R = 0.3 °/s
3. Hold 4°C
Ligation
- left ITR fragment: 6.2 µL
- Suffix: 1 µL
- pGA14: 11.5 µL
- Buffer: 2.2 µL
- T4 Ligase: 1 µL
Total: 21.9 µL
Sample will be incubating over night in the thermocycler at 16°C.
Continuation: right ITR
Investigator: Hanna
Sequencing results delivered that the right ITR was succesfully ligated to the pGA14 plasmid. Unfortunately just about 100 bp were successfully sequenced. Within the ITR region sequencing seemed one more time not to be possible due to the very strong secondary structures which hinder the polymerase to read through. Therefore we can just validate that the ITR with the prefix is inside pGA14, but it's not possible to give a statement about the ITR quality or whether the suffix was successfully fused to the sequence. Theoretically the digested plasmid is not able to religate without ITR and Suffix sequence. But noone knows what our E.colis are able to achieve :)
Nevertheless we decided to continue with the experiment:
Practical Cloning:
Digestion of pGA14_Cerulean (P61) and of pGa14_Präfix-PreSuffix-rITR (P115)
| components | volume of pGA14_Cerulean (P61) /µl | volume of pGa14_Präfix-PreSuffix-rITR (P115) /µl |
| DNA | 7.5 | 7.5 |
| BSA (100x) | 0.2 | 0.2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme 1 | EcoRI-HF: 1 | EcoRI-HF: 1 |
| Enzyme 2 | SpeI: 1 | SpeI: 1 |
| H2O | 8.3 | 8.3 |
| Total volume | 20 | 20 |
- Incubation: ~2 h
Agarose-Gel:
P61:
0.5 g Agarose, 50 mL TBE!!! (1 %), 3 µL GELRED, at 115 Volt, running time: 45 minutes
P115:
0.8 g Agarose, 50 mL TBE!!! (1.45 %), 3 µL GELRED, at 115 Volt, running time: 35 minutes
| Sample | Sample/µl] | Loading dye (5x)/µl | Expected size 1 (Geneious) | Expected size 2 (Geneious) |
|---|---|---|---|---|
| P61 | 20 µl | 5 µl | 2895 bp | ~ 750 bp |
| P115 | 20 µl | 5 µl | 171 bp | ~ 2890 bp |
- Marker: GeneRuler ladder mix
Gelextraction
Gel measurement:
| Sample | Concentration |
| P61 | 16.65 ng/µL |
| P115 | 7.95 ng/µL |
Ligation
- right ITR fragment: 2.2 µL
- pGA14: 5.8 µL
- Buffer: 1 µL
- T4 Ligase: 1 µL
Total: 10 µL
Sample will be incubating over night in the thermocycler at 16°C.
ITR gel pictures
Digestion worked. The marked bands were cut out.
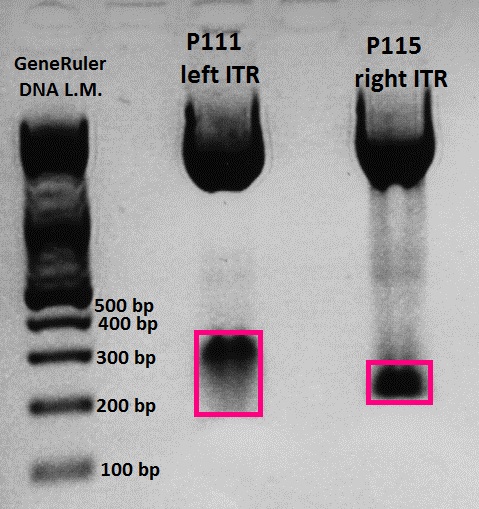
Digestion of the right ITR fragment worked well. Unfortunately the left ITR fragment is too large. The ecpectation was ~ 151 bp. It seemed to consist of ~ 300 bp. --> Two ITRs in tandem??? Hopefully GATC will find a way in order to sequence the ITRs!
ZEGFR:1907
Hanna (guided by Sven)
The Affibody-BioBrick (RFC25) was ordered at Geneart. The sequence does not contain any iGEM restriction sites. We'll receive the DNA in ~ 8 work days.
Strategy: See Lab day 28.07.2010.
More information: See Informationssammlung zum AAV and Theoretical cloning !!
Mini-Prep and Test digestion of pSB1C3_SDM_SspI, pSB1C3_SDM_PvuII, pSB1C3_beta-globin, pSB1C3_hgH, pAAV_RC_1.1_BamHI long PCR, pAAV_RC_1.1_BamHI short PCR,pGL3_hTERT and pAAV_RC_1.1_SalI (short PCR), pAAV_RC_1.1_BamHI long PCR, pAAV_RC_1.1_BamHI short PCR
Investigator: Anissa, Stefan
Comments: There are some (SspI, PvuII, BamHI, SalI) restriction sites in our standard vector which are also present in the viral loops. Therefore, the sites in the vector have to be removed. The beta-globin and hGH constructs are BioBricks for sending to iGEM headquarters. Sequencing is needed. hTERT is tumor-specific promotor we obtained from Kira. Sequencing is needed.
Mini-Prep of pSB1C3_SDM_SspI clone 1 to 4: After performing Mini-Preps concentration was measured with Nanodrop:
- pSB1C3_SDM_SspI clone 1 (=P125): c = 321,9 ng/µl
- pSB1C3_SDM_SspI clone 2 (=P126): c = 290,3 ng/µl
- pSB1C3_SDM_SspI clone 3 (=P127): c = 309,0 ng/µl
- pSB1C3_SDM_SspI clone 4 (=P128): c = 298,1 ng/µl
Mini-Prep of pSB1C3_SDM_PvuII clone 1 to 4:
After performing Mini-Preps concentration was measured with Nanodrop:
- pSB1C3_SDM_PvuII clone 1 (=P129): c = 308,4 ng/µl
- pSB1C3_SDM_PvuII clone 2 (=P130): c = 316,9 ng/µl
- pSB1C3_SDM_PvuII clone 3 (=P131): c = 445,6 ng/µl
- pSB1C3_SDM_PvuII clone 4 (=P132): c = 445,4 ng/µl
Mini-Prep of pSB1C3_beta-globin clone 1 and 2:
After performing Mini-Preps concentration was measured with Nanodrop:
- pSB1C3_beta-globin clone 1 (=P133): c = 221,9 ng/µl
- pSB1C3_beta-globin clone 2 (=P134): c = 206,3 ng/µl
Mini-Prep of pSB1C3_hgH clone 1 and 2:
After performing Mini-Preps concentration was measured with Nanodrop:
- pSB1C3_hgH clone 1 (=P135): c = 245,5 ng/µl
- pSB1C3_hgH clone 2 (=P136): c = 226,2 ng/µl
Mini-Prep of pAAV_RC_1.1_BamHI (long PCR) clone 1 and 2:
After performing Mini-Preps concentration was measured with Nanodrop:
- pAAV_RC_1.1_BamHI (long PCR) clone 1 (=P137): c = 537,8 ng/µl
- pAAV_RC_1.1_BamHI (long PCR) clone 2 (=P138): c = 546,9 ng/µl
Mini-Prep of pAAV_RC_1.1_BamHI (short PCR) clone 1 and 2:
After performing Mini-Preps concentration was measured with Nanodrop:
- pAAV_RC_1.1_BamHI (short PCR) clone 1 (=P139): c = 547,0 ng/µl
- pAAV_RC_1.1_BamHI (short PCR) clone 2 (=P140): c = 494,0 ng/µl
Mini-Prep of pGL3_hTERT clone 1 and 2:
After performing Mini-Preps concentration was measured with Nanodrop:
- pGL3_hTERT clone 1 (=P141): c = 286,1 ng/µl
- pGL3_hTERT clone 2 (=P142): c = 319,9 ng/µl
Mini-Prep of pAAV_RC_1.1_SalI (short PCR) clone 1 and 2:
After performing Mini-Preps concentration was measured with Nanodrop:
- pAAV_RC_1.1_SalI (short PCR) clone 1 (=P143): c = 480,0 ng/µl
- pAAV_RC_1.1_SalI (short PCR) clone 2 (=P144): c = 510,7 ng/µl
Test digestion:
pSB1C3_SDM_SspI was digested with SspI.
pSB1C3_SDM_PvuII was digested with PvuII.
pSB1C3_beta-globin was digested with XbaI and PstI.
pSB1C3_hgH was digested with XbaI and PstI.
pAAV_RC_1.1_BamHI (long PCR) was digested with BamHI.
pAAV_RC_1.1_BamHI (short PCR) was digested with BamHI.
pGL3_hTERT was digested with BglII and MluI.
pAAV_RC_1.1_SalI (short PCR) was digested with SalI.
| components | volume of P125/µl | volume of P126/µl | volume of P127/µl | volume of P128/µl | volume of neg. control P51.1/µl | volume of P129/µl | volume of P130/µl | volume of P131/µl | volume of P132/µl | volume of neg. control P51.1/µl | volume of P133/µl | volume of P134/µl | volume of P135/µl | volume of P136/µl | volume of P137/µl | volume of P138/µl | volume of neg. control P53.3/µl | volume of P139/µl | volume of P140/µl | volume of neg. control P53.2/µl | volume of P141/µl | volume of P142/µl | volume of P143/µl | volume of P144/µl | volume of neg. control P53.1/µl |
| DNA | 3 | 3 | 3 | 3 | 7 | 3 | 3 | 2 | 2 | 7 | 4 | 4 | 4 | 4 | 1,5 | 1,5 | 2 | 1,5 | 2 | 2 | 3 | 3 | 2 | 1,8 | 2 |
| BSA (10x) | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 |
| Buffer 4 (10x) | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 | 1,5 |
| Enzyme 1 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 |
| Enzyme 2 | / | / | / | / | / | / | / | / | / | / | 0,5 | 0,5 | 0,5 | 0,5 | / | / | / | / | / | / | 0,5 | 0,5 | / | / | / |
| H2O | 8,5 | 8,5 | 9,5 | 9,5 | 4,5 | 8,5 | 8,5 | 8,5 | 8,5 | 4,5 | 7 | 7 | 7 | 7 | 10 | 10 | 9,5 | 10 | 9,5 | 9,5 | 8 | 8 | 9,5 | 9,7 | 9,5 |
| Total volume /µl | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 |
Expected sizes:
pSB1C3_SDM_SspI
- no cut
control:
- 2072bp
pSB1C3_SDM_PvuII
- no cut
control:
- 2072bp
pSB1C3_beta-globin
- 756bp
- 2058bp
pSB1C3_hgH
- 502bp
- 2058bp
pAAV_RC_1.1_BamHI (long and short PCR):
- no cut
control:
- 811bp
- 6524bp
pGL3_hTERT
- 467bp
- 4801bp
pAAV_RC_1.1_SalI (short PCR):
- no cut
control:
- 7331bp
Results of samples loaded 1% agarose gel and ran ~50 minutes at 115 V:
Sequencing
Sequencing will be done next week.
Midi-prep of pSB1C3_CFP
Investigator: Anissa
Comments: The mini-prep of pSB1C3_CFP was depleted, so a double midi-prep was made for serving the pool. They were named p51.1 and p51.2. Both are from the glycerol-stock B39, which means they are sequenzed and ready to use.
- concentration p51.1: 132,06 ng/µL
- concentration p51.2: 151,47
75. Labortag 31.7.2010
Trafo of ITR-over-night ligations
Investigator: Hanna
The last step of the ITR fancy method was done ytesterday. Means: If cloning worked, both ITRs should be in the RFC10 standard. Ligation was done over night and the referring trafo will be done today.
Trafo was performed following the standard protocol.
Abberations:
3 µL sample was added to BL21 cells. Incubation time on ice was just 15 minutes. Incubation at 37°C for 45 minutes.
 "
"