Team:Freiburg Bioware/NoteBook/Labjournal/September2
From 2010.igem.org
(→Contuniation: Cloning pSB1C3_Viral Brick 587KO-empty (B274/P542) into pSB1C3_VP2/3_Capins (B438/P514)) |
(→Contuniation: Cloning pSB1C3_Viral Brick 587KO-empty (B274/P542) into pSB1C3_VP2/3_Capins (B438/P514)) |
||
| Line 1,788: | Line 1,788: | ||
pSB1C3_VP2/3_Capins_587KO-empty clone 3: 23 µl Buffer 4, 0,5 µl BamHI, 0,5 µl PvuII, 20 µl DNA, 6 µl H2O <br> | pSB1C3_VP2/3_Capins_587KO-empty clone 3: 23 µl Buffer 4, 0,5 µl BamHI, 0,5 µl PvuII, 20 µl DNA, 6 µl H2O <br> | ||
Because somebody switched the 37°C thermoblock off i dont know the exact digestion time. Therefore i switched the thermoblock on again and waited 1 hour. I used that much DNA because the desired insert is 48 bp long. | Because somebody switched the 37°C thermoblock off i dont know the exact digestion time. Therefore i switched the thermoblock on again and waited 1 hour. I used that much DNA because the desired insert is 48 bp long. | ||
| - | [[Image:Freiburg10_0924_pat.jpg]] | + | [[Image:Freiburg10_0924_pat.jpg]] |
| - | + | ||
Clone 3 was sent for sequencing. | Clone 3 was sent for sequencing. | ||
Revision as of 15:56, 24 September 2010

September
124. labday 19.09.2010
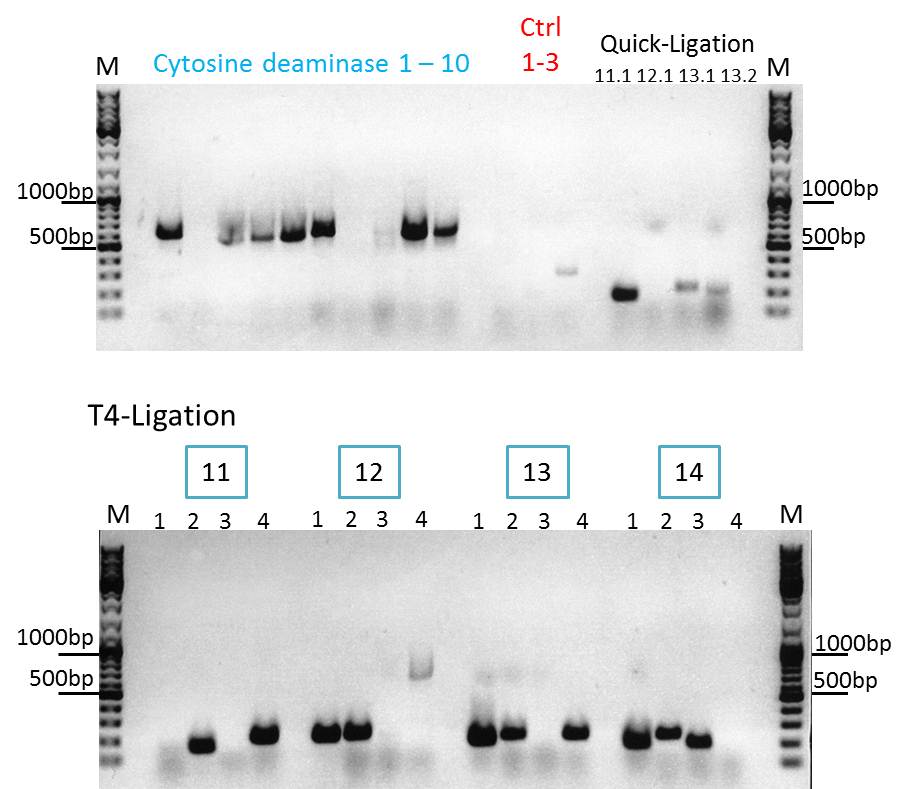
Continuation with Colony-PCR of pSB1C3_CD and ViralBrick motif Z34C
Investigator: Bea
Comment: Since the colony PCR of the last try did not work exactly the way we expected it, another colony PCR approach with new primers was conducted.
Loading plan:
upper lanes
M CD1 CD2 CD3 CD4 CD5 CD6 CD7 CD8 CD9 CD10 +ctrl(CFP) +ctrl(BAP) -ctrl(pAAV_MCS) 11Q1 12Q1 13Q1 13Q2 M
lower lanes
M 11T41 11T42 11T43 11T44 12T41 12T42 12T43 12T44 13T41 13T42 13T43 13T44 14T41 14T42 14T43 14T44 M
Expected sizes of the PCR products are:
- Cytosine deaminase (CD) = 1300bp
- Z34C loops insertion motif = 220bp
- Positive control 1 = CFP = 750bp
- Positive control 2 = BAP_587 = 170bp
Results:
The Colony PCR of the Cytosine Deaminase (CD) did not work out. The detectable bands at around 700 bp correspond to CFP. Therefore, BioBrick production of the CD must be repeated. The following three bands represent the controls. Three different controls were used. The first corresponds to pSB1C3_CFP, the second to BAP_587 and third control is the negative control. Unfortunately now bands can be detected in the postive controls. A faint band can be seen in the negative control which is the only lane in which no band should be detectable.
The following bands after the three controls correspond to the approaches of the Z34C Viralbrick production. Some bands in the lower and the upper lanes show positive results. The corresponding inoculated overnight culture were centrifuged and prepared in order to perform a Mini-Prep tomorrow.
Comment: hmm, seems that the CD-assambly didn't work at all.. Should i try it again or are we gonna skip it for good due to other priorities?(kira)
No, we cannot skip it. Can we change something else?? Any ideas what we can alter in the protocol? Because problem is that we are still waiting for the tk... so we should aswell focus on the other prodrug activating enzmye!! (Bea)
Testtransduction of the final modified capsid coding constructs
Investigator: Adrian
Aim of the experiment:
During the project 22 silent nucleotide exchange mutations were introduced into the capsid coding construct to make it compatible with the RFC standards and to have single cutting restriction enzymes flanking the 453 and the 587 loop sequence. Two point mutations had to be dismissed, because either a first test transduction showed that the construct was not working anymore or the insertion of the synthesized gene posed serious problems, because the restriction enzyme did not work.
Finally, we have a construct that is shown to produce infectious particles comparable to the current AAV systems and carries 20 point mutations. Now we can announce that the Adeno-associated Virus is compatible to the RFC standard and the idea to replace the loop sequences via ViralBricks works!
Impression from the moment:
Miniprep of serveral constructs
Investigator: Stefan
Glycerol stocks were prepared:
- B430 = pCerulean_ZEGFR:1907_Longlinker_VP2/3
- B431 = pCerulean_CFP_Middlelinker_VP2/3_insCap
- B432 = pCerulean_ZEGFR:1907_Shortlinker_VP2/3_insCap
Mini-Prep was performed according to the standard protocol
- P527 = pCerulean_ZEGFR:1907_Longlinker_VP2/3 c = 230,61 ng/µl
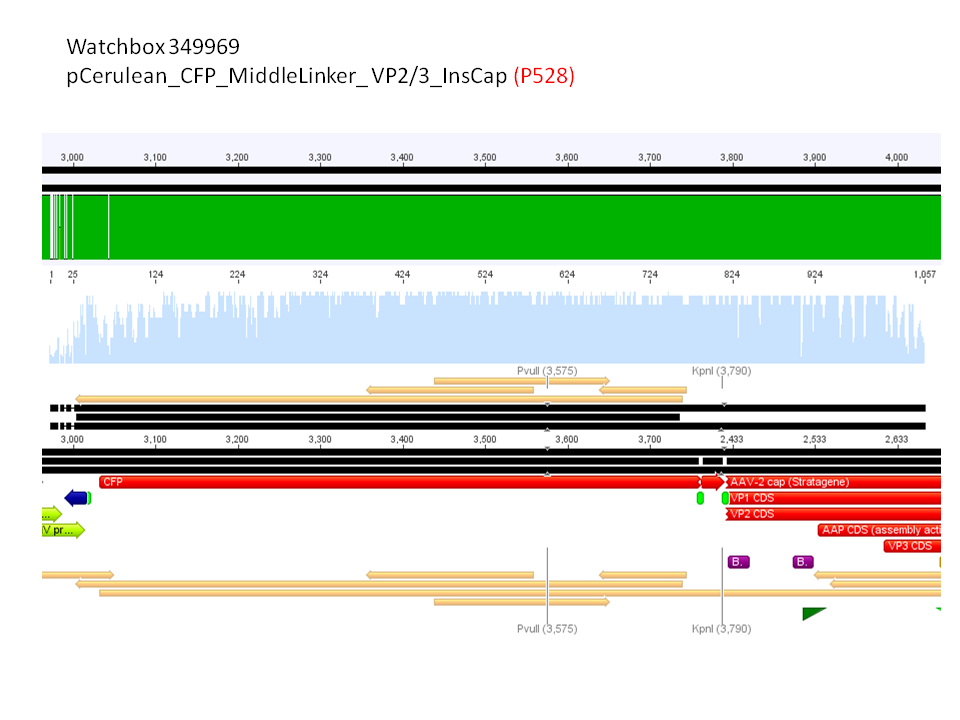
- P528 = pCerulean_CFP_Middlelinker_VP2/3_insCap c = 290,81 ng/µl
- P529 = pCerulean_ZEGFR:1907_Shortlinker_VP2/3_insCap c = 269,39 ng/µl
Several other constructs were preped, but need to be confirmed before being added to plasmid/glycerol stock.
125. labday 20.09.2010
Picking the clones and preparing the over-night culture
Investigator: Kira
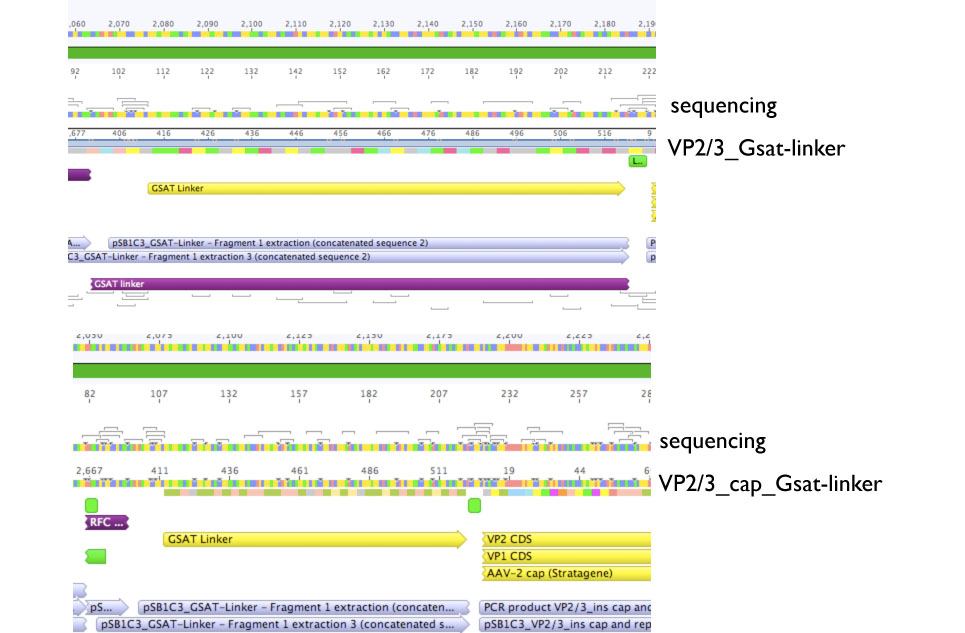
VP2/3_Gsat-linker as well as VP2/3_cap_Gsat-linker plates contain colonies, thus 3 clones from each plate were inoculated into 5 ml DYT+ 5ul Chloramphenicol and incubated @ 37 C
Minipreps and Sequencing of ViralBrick clones
Investigator: Achim
The following clones were prepped:
- Quickligation:
- 11.1, c = 85,41 ng/µl
- 13.1, c = 96,93 ng/µl
- T4-Ligation:
- 11.4, c = 76,42 ng/µl
- 12.1, c = 80,36 ng/µl
- 12.4, c = 85,43 ng/µl
- 13.1, c = 90,06 ng/µl
- 13.2, c = 80,87 ng/µl
- 13.4, c = 76,62 ng/µl
- 14.1, c = 76,50 ng/µl
- 14.2, c = 98,52 ng/µl
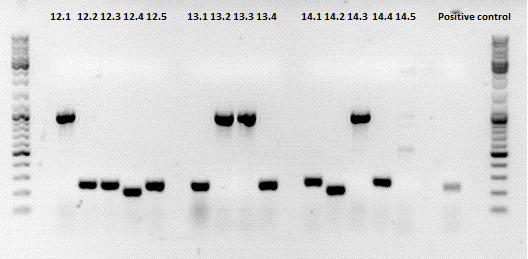
Clones 11.4, 12.1, 13.2, 14.2 (T4) were sent for sequencing.
Cloning of pCerulean_Affibody_VP2/3 and pSB1C3_Affibody_middlelinker_GFP_his
Investigator: Jessica
Comment:Affibody_VP2/3 will be cloned in pCerulean and Affibody_middlelinker will be cloned together with GFP_His in pSB1C3
- P407= pCerulean_CFP_middlelinker c=482,76 ng/µl
- P516= pSB1C3_ZEGFR:1907_VP2/3 clone 1 c= 191,6 ng/µl
- P290= pSB1C3_Affibody_Middlelinker clone 1 c= 227,4 ng/µl
- P518= pSB1C3_GFP_RFC25_upper band clone 1 c= 138,3 ng/µl
- P520= pSB1C3_GFP_RFC25_lower band clone 1 c= 141,6 ng/µl
Digestion:
| components | P407 | P516 | P290 | P518 | P520 |
| DNA | 3,5 | 10 | 7 | 10 | 10 |
| BSA (10x) | 2 | 2 | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 | 2 | 2 |
| EcoRI | 0,8 | 0,8 | - | - | - |
| AgeI | 0,8 | 0,8 | 0,8 | - | - |
| NgoMIV | - | - | - | 0,8 | 0,8 |
| SpeI | - | - | 0,8 | 0,8 | 0,8 |
| H2O | 10,9 | 4,4 | 7,4 | 4,4 | 4,4 |
| Total volume | 20 | 20 | 20 | 20 | 20 |
1,0 g Agarose,100 ml TAE (1%), 6 µl GELRED , at 120 Volt
P516, P518, P520 will be repeated tomorrow (no DNA after gel-ex)

Gelextraction:
The gelextraction was performed according to the standard protocol. DNA concentration of the extracts:
- P407 c= 6,5 ng/µl
- P516 c= - ng/µl
- P290 c= 7,3 ng/µl
- P518 c= - ng/µl
- P520 c= - ng/µl
stands in 4°C room for comtinuation tomorrow
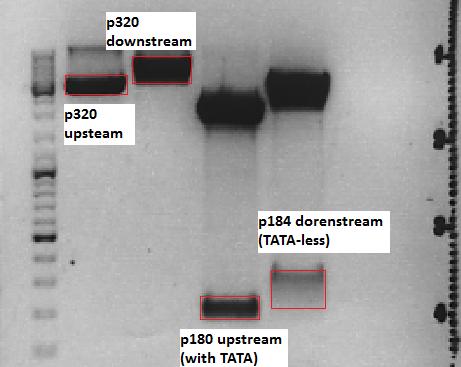
Cloning of P5_TATAless downstream of pSB1C3_001_RC_insrepcap_KpnI_back and P5 upstream of pSB1C3_001_RC_insrepcap_KpnI_back
Investigator: Anissa
Digestion of the constructs:
- P320 = pSB1C3_001_RC_insrepcap_KpnI_back c=408 ng/µL
- P320 = pSB1C3_001_RC_insrepcap_KpnI_back c=408 ng/µL
- P180 = pSB1C3_pTAV2 (P5)clone 1 c=108,8 ng/µL
- P184 = pSB1C3_pAAV_RC (P5TATAless)clone 3 c=176,9 ng/µL
| Components | vP320 upstream /µL | vP180 upstream/µL | vP320 downsteam /µL | vP184 downsteam/µL |
| DNA | 2,5 | 13,8 | 2,5 | 8,5 |
| BSA (10x) | 1,5 | 2 | 1,5 | 1,5 |
| Buffer no. 4 (10x) | 1,5 | 2 | 1,5 | 1,5 |
| Enzyme 1 | XbaI 1 | SpeI 1 | SpeI 1 | XbaI 1 |
| Enzyme 2 | EcoRI 1 | EcoRI 1 | PstI 1 | PstI 1 |
| H2O | 7,5 | 0,2 | 7,5 | 1,5 |
| Total volume | 15 | 20 | 15 | 15 |
Loading plan:
M P320(upstream) P320(downstream) P180 P184
Results:
After gel extraction has been performed, the ligation was carried out.
Ligation:
- v=P320_up =7,37µL
- v=P180_up =0,63µL
- v=P320_down =7µL
- v=P184_down =1µL
The ligation mix was transformed into XL1-Blue cells and plated on agar plates containing chloramphenicol.
Next steps:
Picking clones and perform Mini-Prep.
Test digestion of VP2-N-terminal fusion constructs
Investigator: Hanna
Comment: On Satureday a colony PCR was performed of:
1. pCerulean_Zegfr:1907_ShortLinker_VP2/3 (P530)
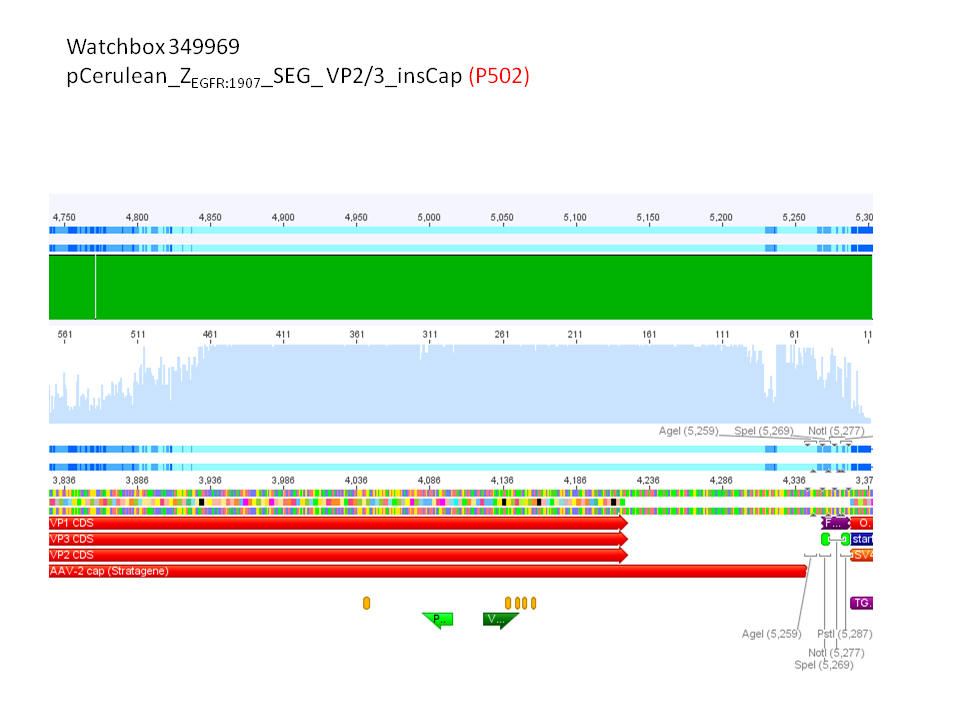
2. pCerulean_Zegfr:1907_SEG_VP2/3 (P531)
3. pCerulean_CFP_MiddleLinker_VP2/3 (P532)
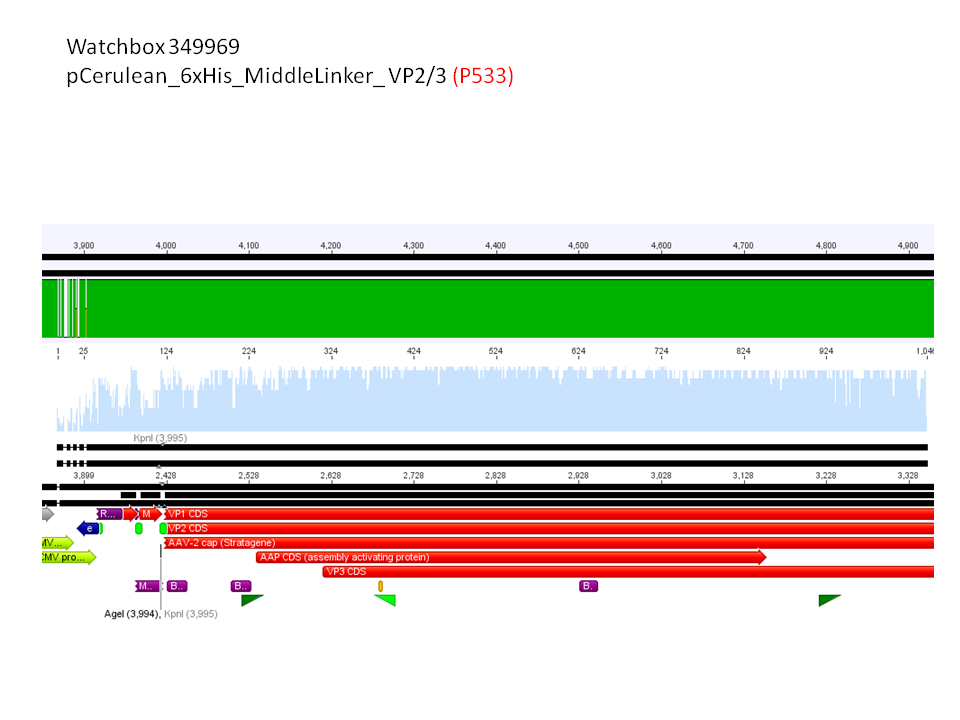
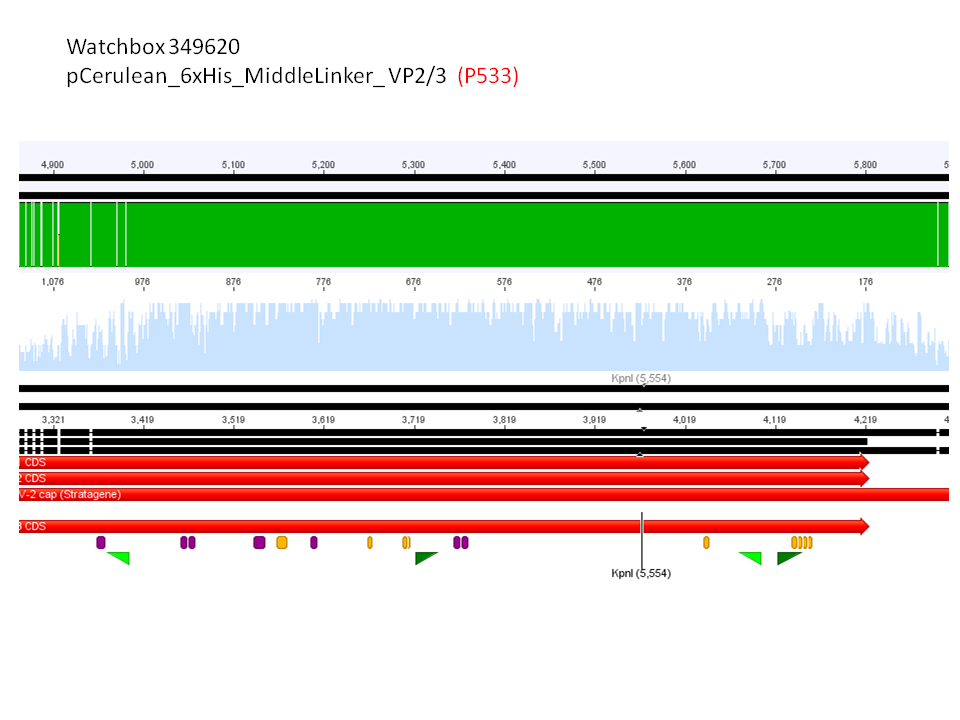
4. pCerulean_6xHis_MiddleLinker_VP2/3 (P533)
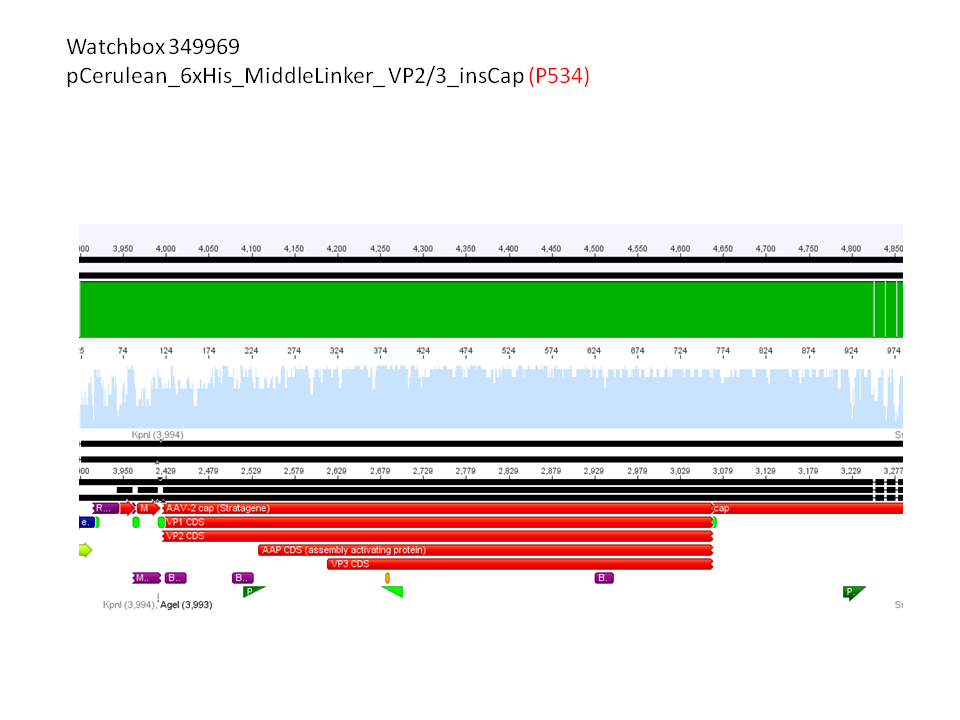
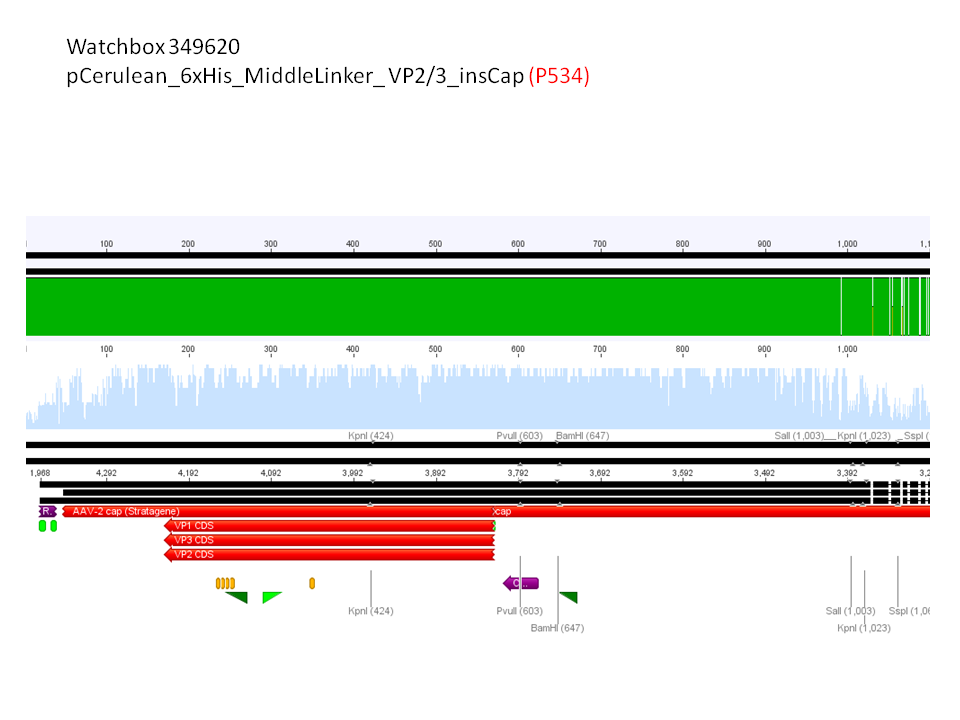
5. pCerulean_6xHis_MiddleLinker_VP2/3_insCap (P534)
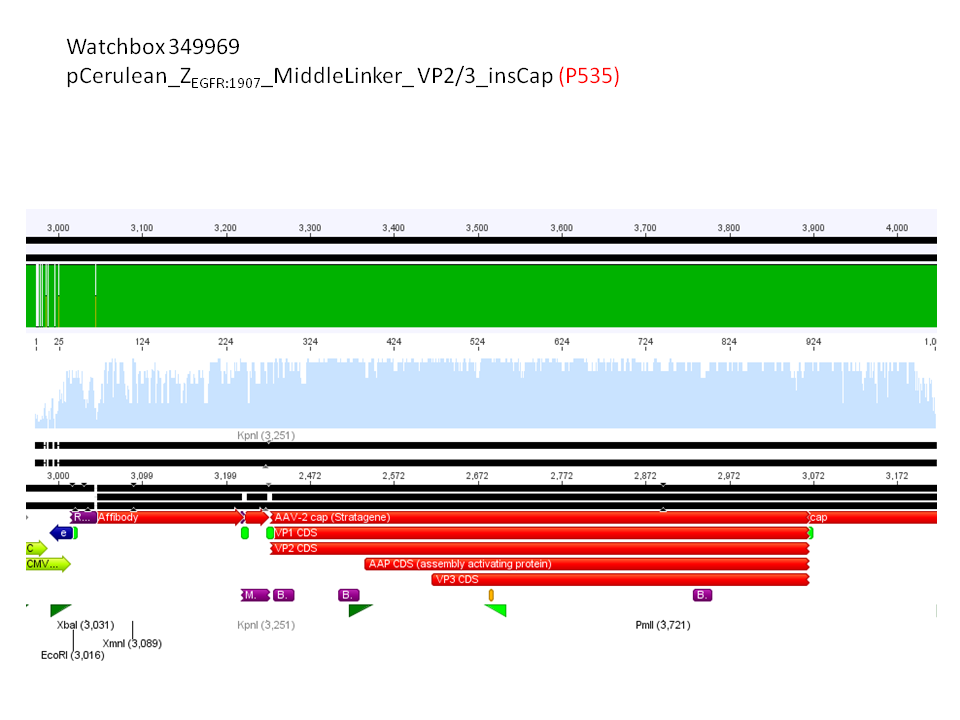
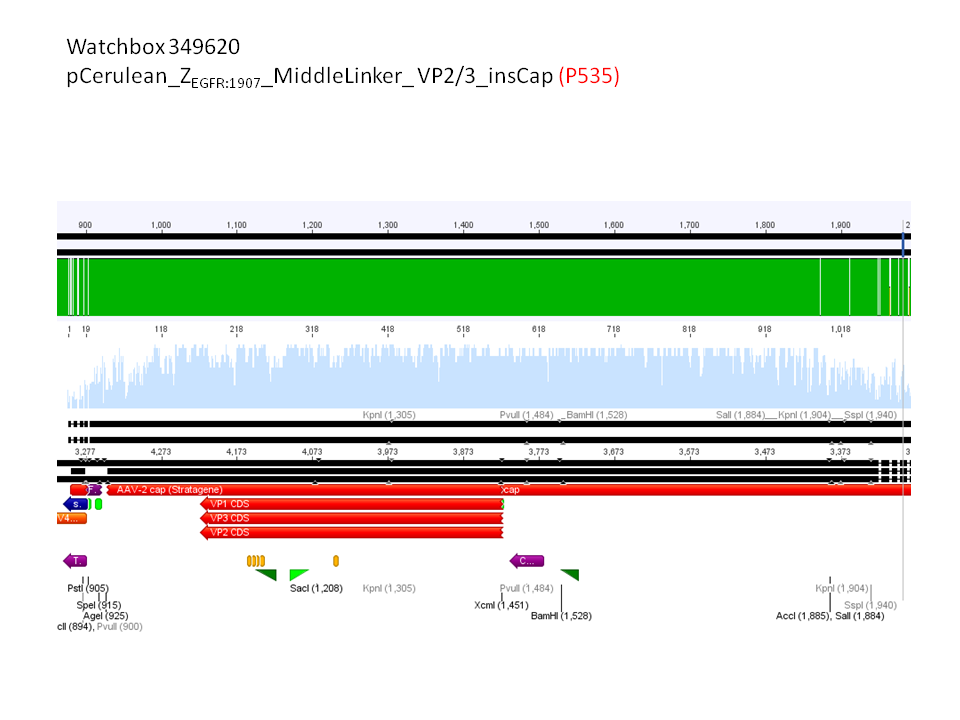
6. pCerulean_Zegfr:1907_MiddleLinker_VP2/3_insCap (P535)
Because all constructs (4 clones of each approach), including the negative control (pAAV_RFC25), showed positive results, one test digestion of each construct was performed.
Digestion
| components | Components Master Mix /µl |
| BSA (10x) | 7 |
| Buffer 4 (10x) | 7 |
| EcoRI | 3.5 |
| SpeI | 3.5 |
| H2O | 35 |
--> 2 µL DNA + 8 µL Master Mix
- Incubation: 1.5 h
Agarose-Gel:
0.4 g Agarose, 50 mL TAE (0.8 %), 3 µL GELRED, at 100 Volt, running time: 50 minutes
| Sample | Sample/µl] | Loading dye (6x)/µl | Expected size 1 | Expected size 2 |
|---|---|---|---|---|
| P 530 | 10 µl | 2 µl | 2710 bp | 3937 bp |
| P 531 | 10 µl | 2 µl | 2710 bp | 3937 bp |
| P 532 | 10 µl | 2 µl | 2710 bp | 3937 bp |
| P 533 | 10 µl | 2 µl | 2710 bp | 3937 bp |
| P 534 | 10 µl | 2 µl | 2710 bp | 3937 bp |
| P 535 | 10 µl | 2 µl | 2710 bp | 3937 bp |
- Marker: GeneRuler ladder mix
| Marker /µL | Sample P530 /µl | Sample P531 /µl | Sample P532 /µl | Sample P533 /µl | Sample P534 /µl | Sample P535 /µl | |
|---|---|---|---|---|---|---|---|
| Lane | 4 | 12 | 12 | 12 | 12 | 12 | 12 |
Comment: Test digestion delivered positive results: Expected fragment size after VP2/3 fusion to Zegfr:1907_Linker: 2180 bp - compared to failed VP2/3 insertion: ~ 220 bp.
Expected fragment size after VP2/3 fusion to CFP_MiddleLinker: 2710 bp - compared to failed VP2/3 insertion: ~ 760 bp.
pCerulean_ZEGFR:1907_Longlinker_VP2/3
pCerulean_CFP_Middlelinker_VP2/3_insCap
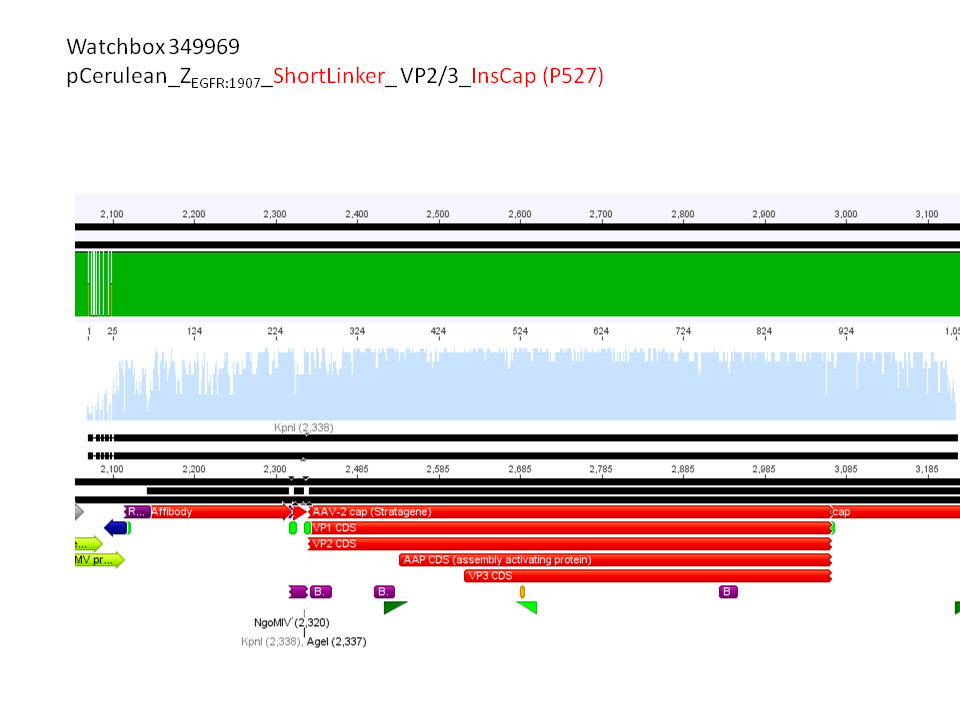
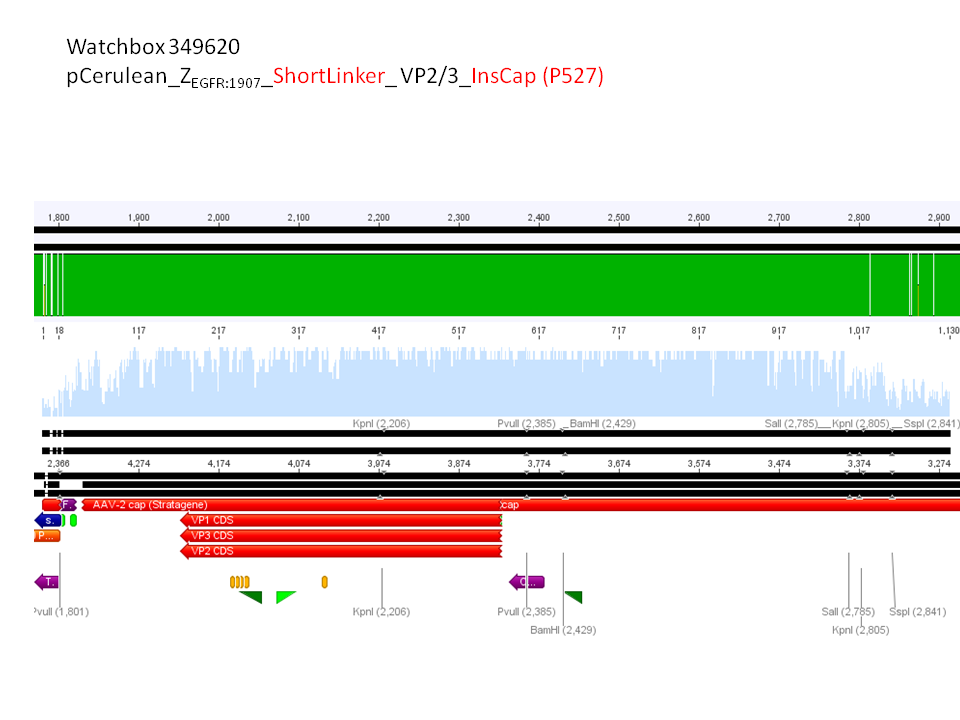
pCerulean_ZEGFR:1907_Shortlinker_VP2/3_insCap
pCerulean_ZEGFR:1907_Shortlinker_VP2/3
pCerulean_ZEGFR:1907_SEG_VP2/3
pCerulean_CFP_MiddleLinker_VP2/3
pCerulean_6xHis_MiddleLinker_VP2/3
pCerulean_6xHis_MiddleLinker_VP2/3_insCap
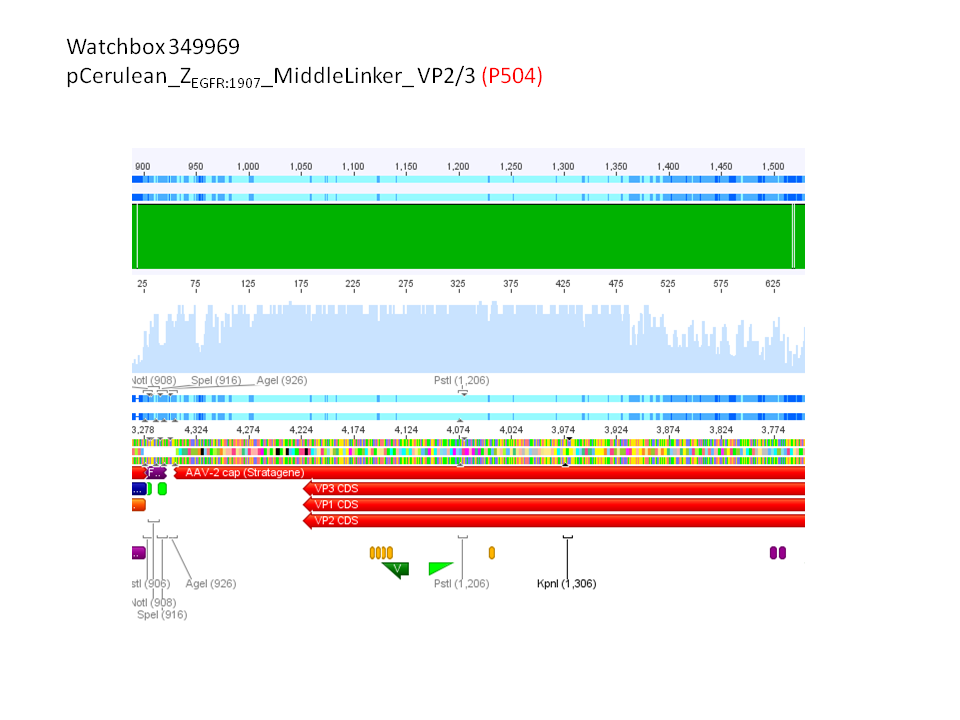
pCerulean_ZEGFR:1907_MiddleLinker_VP2/3_insCap
will be sent for sequencing to GATC.
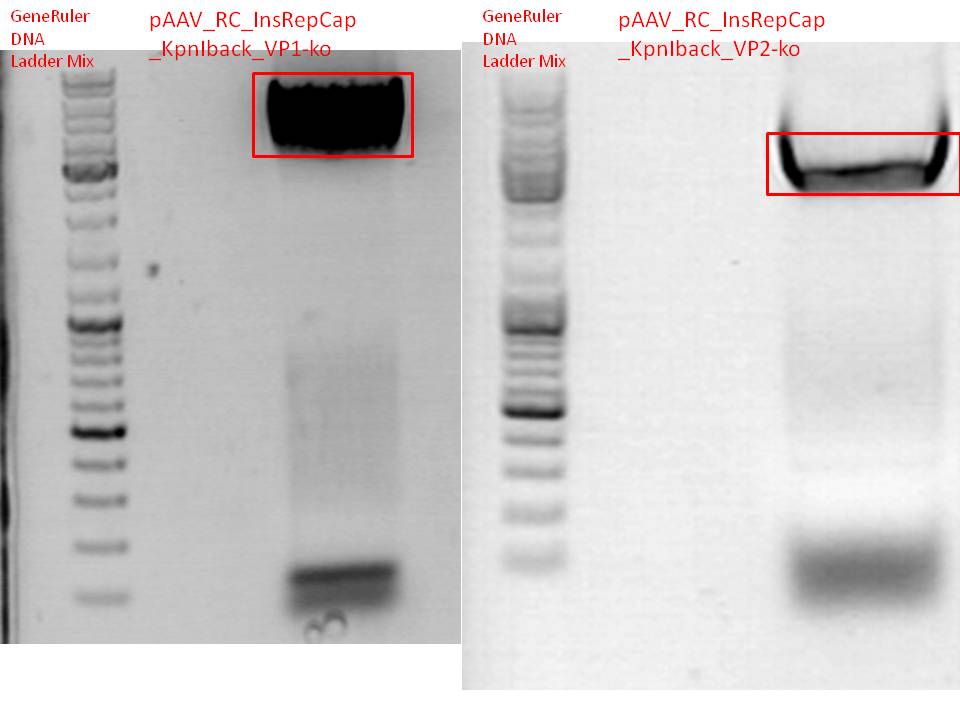
BioBrick production: PCR of pAAV_RC_InsRepCap_KpnIback_VP1-ko and VP2-ko
Investigator: Stefan
DNA samples were diluted 1:1000
- P455 = pAAV_RC_1.2 SDM SalI
- P463 = pAAV_RC_CapIns_prepSDM clone 3
PCR progam:
| Ingredients | Volume P455 / µl | Volume P463 / µl |
| 5X Phusion HF buffer | 10 | 10 |
| 10 mM dNTP mix | 1 | 1 |
| forward primer: O93 | 2,5 | 2,5 |
| reverse primer: O115 | 2,5 | 2,5 |
| DNA Template | 3 | 3 |
| DMSO | 2 | 2 |
| Phusion Polymerase | 0,5 | 0,5 |
| H2O | 28,5 | 28,5 |
| Total volume | 50 | 50 |
PCR program:
| Cycles | Temperature / °C | Time / s |
| 1 | 98 | 60 |
| 2 ( step 2-4: 8x) | 98 | 15 |
| 3 | 59 | 25 |
| 4 | 72 | 75 |
| 5 (step 5-6: 17x) | 98 | 15 |
| 6 | 72 | 85 |
| 7 | 72 | 300 |
| Hold | 4 | 4 |
Gel:
0,5 g agarose (1%), 50 ml TAE, 3µl GELRED running time: 45 minutes at 110V
Gel-Extraction:
Gel-extraction was performed according to standard protocol. The complete amount eluted was used for PCR digestion.
Digestion of PCR product:
| Components | PCR products Volume/µL |
| DNA | 30 |
| BSA (10x) | 4 |
| Buffer no. 4 (10x) | 4 |
| EcoRI | 1 |
| SpeI | 1 |
| H2O | - |
| Total volume | 40 |
Digestion was performed at 37 °C for 2 hours.
PCR-Purification:
Gel-extraction was performed according to standard protocol.
- c (P455) = 13,5 ng/µl
- c (P463) = ng/µl
Digestion of plasmid backbone:
Plasmid used: pSB1C3_VCK_Bla (P320)
c (pSB1C3_VCK_Bla) = 408,0 ng/ µl
| Components | vector Volume/µL |
| DNA | 3 |
| BSA (10x) | 2 |
| Buffer no. 4 (10x) | 2 |
| EcoRI | 1 |
| SpeI | 1 |
| H2O | 11 |
| Total volume | 20 |
Digestion was performed at 37 °C for 2 hours.
Gel:
0,5 g agarose (1%), 50 ml TAE, 3µl GELRED running time: 50 minutes at 120V
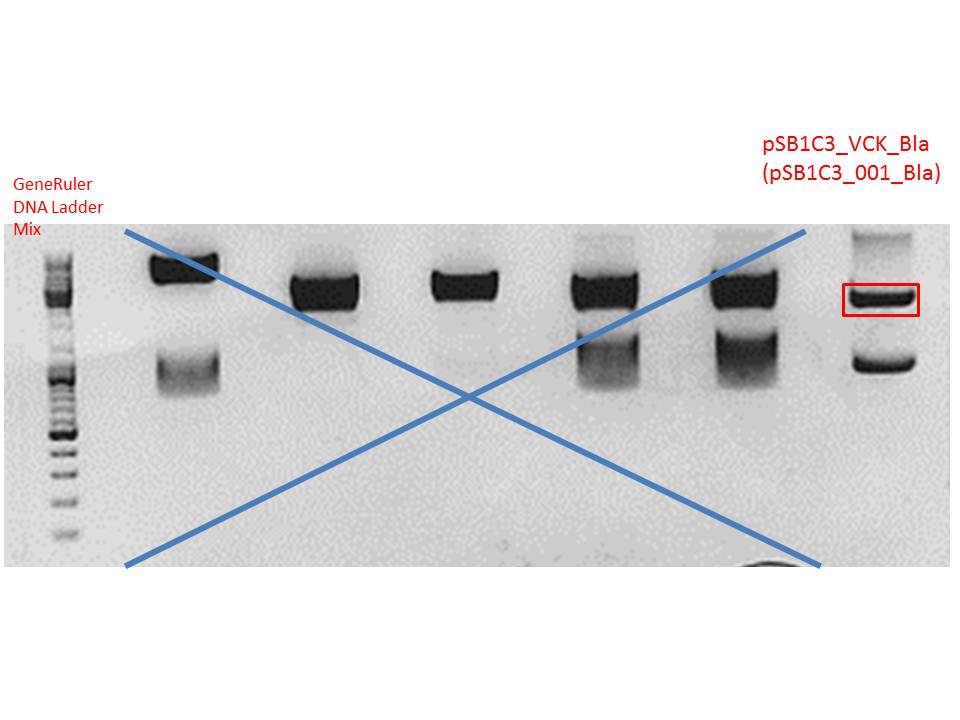
Gel-Extraction:
Gel-extraction was performed according to standard protocol.
c(P320) = 0,8 ng/µl
Ligation:
- 1 µl T4 DNA ligase
- 1 µl 10x Buffer
- 8 µl DNA mix (vector + insert)
Amounts of DNA used:
- c(P320) = 7,73 µl
- c(P455) = 0,27 µl
- c(P320) = 7,19 µl
- c(P463) = 0,81 µl
Incubation time: 45 minutes at room temperature.
Transformation:
Transformation was performed according to standard protocol using XL1b cells and Chloramphenicol as antibiotic.
2x repetition of CD biobrick
Investigator: Kira
| Ingredients | CD sample |
| 5X Phusion HF buffer | 10 µl |
| 10 mM dNTP mix | 1µl |
| forward primer: O158 | 2,5µl |
| reverse primer: O159 | 2,5 µl |
| DNA Template (1:100 dil) | 0,5 µl |
| DMSO | 0 µl |
| Phusion Polymerase | 0,5 µl |
| H2O | 33µl |
| Total volume | 50 µl |
PCR program:
| Cycles | Temperature | Time |
| 98°C | 1 | |
| 10x | 98°C | 15" |
| 58°C | 25" | |
| 72°C | 40" | |
| 17x | 98°C | 15" |
| 65°C | 25" | |
| 72°C | 40" | |
| 1x | 72°C | 5' |
| Hold 4°C |
Digestion of plasmid backbone:
c (pSB1C3) = 151, 1 ng/ µl
| Components | vector Volume/µL |
| DNA 1 µg | 6,0 µl |
| BSA (100x) | 0,2 µl |
| Buffer no. 4 (10x) | 2,0 µl |
| Enzyme 1 XbaI | 0,5 µl |
| Enzyme 2 AgeI HF | 0,5 µl |
| H2O | 10,8 µl |
| Total volume | 20 |
incubation @ 37 C for approx. 6 h
1% agarose gel
According to the gel results, PCR was repeated twice.. But both tries revealed the same unexpected results. Insert from the last cloning procedure was found and used for the ligation and further transformation.
Ligation
T4 ligase was used
1 ul T4 Buffer
1 ul T4 Ligase
8 ul (0,9 ul vector+ 7,1 ul insert) DNA-mix
incubation @ RT for 45 min
Transformation was performed according to the standard protocol and the cells were spread on the agar plate containing Chloramphenicol.
126. labday 21.09.2010
Trafo evalutation of CD biobrick plate
Investigator: Kira
the agar plate was checked at approx. 4 pm and there were no colonies visible. Taking in account that the plate was incubated from 10.30 pm till 8 am @ 37C due to the electricity failure, the plate was put back @ 37C at 4.30 pm
Comment: the plate was picked this morning by a lab member and put in the coldroom
Continuation of cloning of pCerulean_Affibody_VP2/3 and pSB1C3_Affibody_middlelinker_GFP_his
Investigator: Jessica
Comment:Affibody_VP2/3 will be cloned in pCerulean and Affibody_middlelinker will be cloned together with GFP_His in pSB1C3
- P407= pCerulean_CFP_middlelinker c=482,76 ng/µl
- P516= pSB1C3_ZEGFR:1907_VP2/3 clone 1 c= 191,6 ng/µl
- P290= pSB1C3_Affibody_Middlelinker clone 1 c= 227,4 ng/µl
- P518= pSB1C3_GFP_RFC25_upper band clone 1 c= 138,3 ng/µl
- P520= pSB1C3_GFP_RFC25_lower band clone 1 c= 141,6 ng/µl
Digestion:
| components | P516 | P518 | P520 |
| DNA | 10 | 10 | 10 |
| BSA (10x) | 2 | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 | 2 |
| EcoRI | 0,8 | - | - |
| AgeI | 0,8 | - | - |
| MscI | 0,8 | - | - |
| NgoMIV | - | 0,8 | 0,8 |
| SpeI | - | 0,8 | 0,8 |
| H2O | 3,6 | 4,4 | 4,4 |
| Total volume | 20 | 20 | 20 |
0,5 g Agarose,50 ml TAE (1%), 3 µl GELRED , at 120 Volt
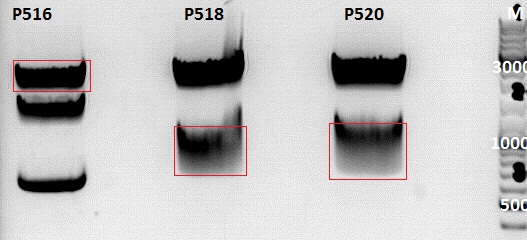
Gelextraction:
The gelextraction was performed according to the standard protocol. DNA concentration of the extracts:
- P516= c= ng/µl
- P518= c= ng/µl
- P520= c= ng/µl
T4 Ligation:
The Ligation was performed as following:
- Vector Volume: µl
- Insert Volume: µl
- 1µl T4-Ligase buffer (10x)
- 8µl (Vector + Insert) mix
- 1µl T4-Ligase
Incubating for 40 minutes.
Transformation:
Trafo was performed according to the standard protocol (XL1blue). The cells were plated on a agar plate with Kana/Cm
Sequenzing result of pSB1C3_EGFP_His upper and (!) lower band
Investigator: Jessica
1. pSB1C3_EGFP_His suffix:
2. pSB1C3_EGFP_His prefix:
results look in both plasmids and are used for cloning of pSB1C3_Affibody_middlelinker_GFP_His
Midi-Prep of pHelper & pSB1C3_leftITR_CMV_beta-globin_mVenus_hGH_rightITR
Investigators: Chris W.
Midi-Preps of pHelper=P540 and pSB1C3_leftITR_CMV_beta-globin_mVenus_hGH_rightITR=P541
The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| plasmid-no. | P540 | P541 |
| concentration (ng/µl) | 480,45 | 1264,20 |
New aliquots (60µl) of XL1blue can be used ! (Jessica)
127. labday 22.09.2010
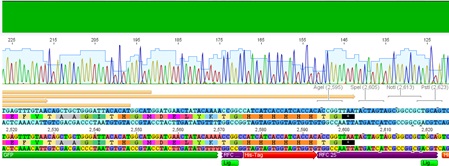
Sequencing results of ViralBrick ligations using dephosphorylated vector
Investigator: Achim
Sequencing confirmed a correct sequence for clone 11.4 (453 Z34C). Clone 12.1 (587 Z34C)has several missing bases at the 3' end of the insert, clone 13.2 has an a -> c mutation and a c insertion in the insert (although the a->c might be a sequencing error.). Clone 14.2 contained a c deletion.
We have several new approaches:
-Correction of the mutations via PCR: We'll use the hybridisation oligo for the side of the insert containing the mutations, the rfc 25 primer on the other side. The amplified insert will be ligated into dephosphorized vector.
-As a backup, SDM-primers will be ordered and should be available next week.
-Also, a new colony pcr with several clones from the last ligation plates will be carried out.
-clone 13.4 (already prepped) will be sent for sequencing
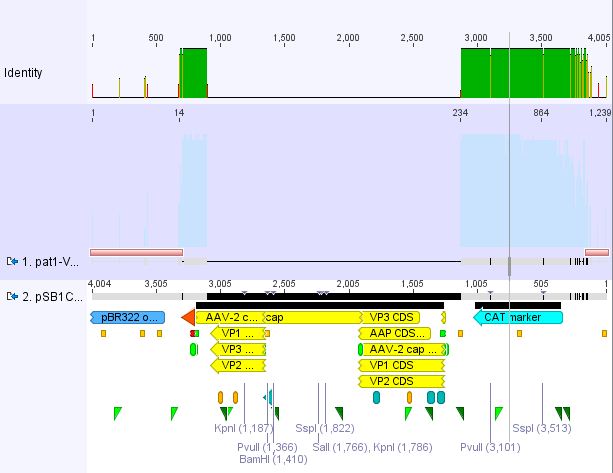
Cloning pSB1C3_Viral Brick 587KO-empty (B274/P542) into pSB1C3_VP2/3_Capins (B438/P514)
Investigator Patrick
After an over night digestion with BamHI and PvuII at 37°C ....
The Gelextraction with P514 was performed according to the standard protocol yielding 16,9 ng/ml.
The P542 insert should be 48 bp long and therefore was extracted with a QIAGEN kit able to extract even very small DNA fragments (40 bp) (QiaEX II Gel Extraction Kit (150)).
Expected size of the fragments:
P542: 2112 & 48 bp
P514: 3956 & 48 bp
The 48 bp band of P514 can't be seen clearly on the picture due to the bad resolution.
Ligation with T4 DNA ligase: 1 µl buffer (10x), 1 µl T4 DNA ligase, 7 µl Vector, 1 µl Insert, 70 minutes.
The transformation with XL1B was performed according to the standard protocol and plated: pSB1C3_VP2/3_Capins_587KO-empty
Cellculture: Serum-free cells
Investigator Patrick
We want our virus production to be serum-free to simplify the following purification.
15 ml cell-medium, 25% serum-free: 3,75 ml serum-free DMEM + 11,25 ml DMEM
15 ml cell-medium, 50% serum-free: 7,5 ml serum-free DMEM + 7,5 DMEM
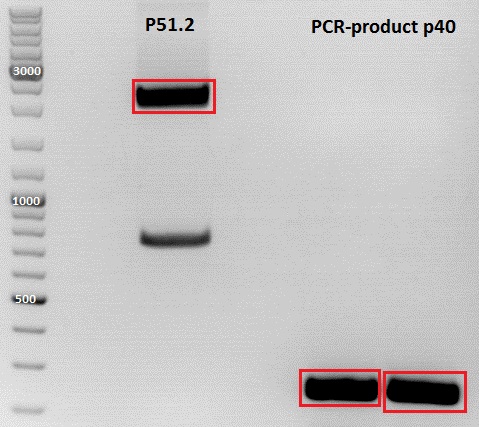
PCR of P40
Investigator: Jessica
DNA sample P432 was diluted 1:1000
| Ingredients | Volume / µl |
| 5X Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| forward primer: O152 | 2,5 |
| reverse primer: O188 | 2,5 |
| DNA Template | 3,0 |
| DMSO | - |
| Phusion Polymerase | 0,5 |
| H2O | 31 |
| Total volume | 50,5 |
PCR program:
| Cycles | Temperature / °C | Time / s |
| 98 | 60 | |
| 8x | 98 | 15 |
| 61 | 25 | |
| 72 | 8 | |
| 17x | 98 | 15 |
| 69 | 25 | |
| 72 | 8 | |
| 1x | 72 | 300 |
| Hold | 4 |
Digestion of plasmid backbone:
Plasmid used: pSB1C3_CFP (P51.2)
c (pSB1C3_CFP) = 151, 1 ng/ µl
| Components | vector Volume/µL |
| DNA | 10 |
| BSA (10x) | 2 |
| Buffer no. 4 (10x) | 2 |
| XbaI | 1 |
| PstI | 1 |
| H2O | 4 |
| Total volume | 20 |
incubation @ 37 C for approx. 2 h
Comment:
Digestion of PCR product:
| Components | PCR products Volume/µL |
| DNA | 40 |
| BSA (10x) | 6 |
| Buffer no. 4 (10x) | 6 |
| PstI | 2 |
| XbaI | 2 |
| H2O | 4 |
| Total volume | 60 |
Digestion was performed at 37 °C for 2 hours.
PCR-Purification:
- c (P432) = 31,8ng/µl
Gel-Extraction:
Gel-extraction was performed according to standard protocol.
c(P51.2) = 3,7ng/µl
Ligation:
- 1 µl T4 DNA ligase
- 1 µl 10x Buffer
- 8 µl DNA mix (vector + insert)
Amounts of DNA used:
- (P51.2) = 7,74 µl
- (P432) = 0,26µl
Incubation time: 45 minutes at room temperature.
Transformation:
Transformation was performed according to standard protocol using XL1b cells and Chloramphenicol as antibiotic.
Mini-Prep and test digestion of pSB1C3_001_RC_InsRepCap_KpnIback_VP1-ko
Investigator: Stefan
Comment:
Glycerol stocks were prepared:
- B463 = pSB1C3_001_RC_InsRepCap_KpnIback_VP1-ko_P5tataless clone 1
- B464 = pSB1C3_001_RC_InsRepCap_KpnIback_VP1-ko_P5tataless clone 2
Mini-Prep was performed according to standard protocol:
- P548 = pSB1C3_001_RC_InsRepCap_KpnIback_VP1-ko_P5tataless clone 1 c = 435,82 ng/µl
- P549 = pSB1C3_001_RC_InsRepCap_KpnIback_VP1-ko_P5tataless clone 2 c = 409,21 ng/µl
Test digestion:
Results:
500px
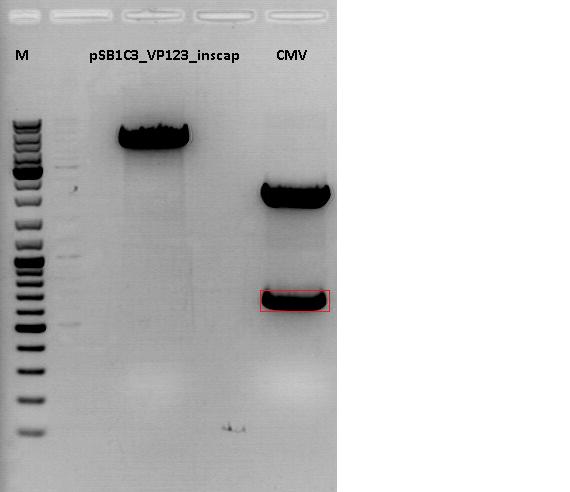
Cloning of CMV promotor into pSB1C3_VP123_inscap
Investigator: Anna
- Vector: name: pSB1C3_VP123_inscap P489
- Insert: name: pSB1C3_CMV P193
- new vector name: pSB1C3_CMV_VP123_inscap P
- buffer used: 4
- DNA concentration (vector): 350,0 ng/µl ; DNA concentration (insert): 333,5 µg/µl
| components | volume of pSB1C3_VP123_inscap /µl | volume of pSB1C3_CMV /µl |
| DNA | 3 | 4,5 |
| BSA (10x) | 1,5 | 1,5 |
| Buffer 4 (10x) | 1,5 | 1,5 |
| Enzyme I | EcoI HF 1 | EcoI HF 1 |
| Enzyme II | XbaI 1 | SpeI 1 |
| H2O | 7 | 5,5 |
| Total volume (e.g. 15,20,25,30 µl) | 15 | 15 |
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 120 Volt, running time:45
Loading plan for agarose gel:
Loading dye with SDS (6X), 3 µl
Marker used: GeneRuler ladder mix (Fermentas), 4 µl
| Marker | Sample 489, 18µl | Sample 193, 18µl | |
|---|---|---|---|
| Lane | 1 | 3 | 5 |
| Fragment size | 4404 bp | 681 bp |
Gel extraction:
| pSB1C3_VP123_inscap_cut | CMV | |
|---|---|---|
| DNA-concentration [ng/µl] | 14,09 | 32,61 |
T4 Ligation:
Volume vector: 3,86 µl
Volume insert: 4,14 µl
Transformation:
XL1B cells were used.
Sequencing of Zegfr:1907-Linker Library, His-Tag and imaging appoaches
Investigator: Hanna
Comment: After several problems were managed (cloning difficulties, colony PCR troubles, interchanges of samples,...), sequences of the final constructs needed to be verified in order to test the constructs in cell culture.
Comment: Sequences looked well. The 6 samples with VP2/3_insCap will be tested today in cell culture.
Mass Midi-Prep
Investigator: Chris W.
Midi-Prep of:
pSB1C3_001_RC_InsRepCap_KpnIback_P5tataless clone 1 =P544
pSB1C3_001_RC_InsRepCap_KpnIback_P5tataless clone 2 =P545
pSB1C3_001_P5_RC_InsRepCap_KpnIback clone 1 =P546
pSB1C3_001_P5_RC_InsRepCap_KpnIback clone 2 =P547
pHelper =P550
pHelper =P551
pSB1C3_litr_pTert_ßglobin_mvenus_hgh_ritr clone 1 =P552
pAAV_RC_RepCapIns_SDMKpnl_VP1ko clone 1 =P553
pAAV_RC_RepCapIns_SDMKpnl_VP2ko clone 1 =P554
pAAV_RC_RepCapIns_SDMKpnl_VP2ko clone 1 =P555
pCerulean_ZEGFR:1907_SEG_VP2/3_CapIns clone 3 =P556
pCerulean_ZEGFP:1907_LongLinker_VP2/3_CapIns clone 2 =P557
pCerulean_CFP_Middlelinker_VP2/3_insCap =P558
pCerulean_ZEGFR:1907_Shortlinker_VP2/3_insCap =P559
pCerulean_6xHis_MiddleLinker_VP2/3_insCap =P560
pCerulean_ZEGFR:1907_MiddleLinker_VP2/3_insCap =P561
pCerlulean_ZEGFR:1907_Shortlinker_VP2/3_insCap clone 4.8 =P562
The Midi-Preps were performed according to the standard protocol yielding the following concentrations:
| plasmid-no. | P544 | P545 | P546 | P547 | P550 | P551 | P552 | P553 | P554 | P555 | P556 | P557 | P558 | P559 | P560 | P561 | P562 |
| concentration (ng/µl) | 1851,91 | 1822,3 | 1783,48 | 2625,57 | 1524,63 | 3333,48 | 2641,57 | 2165,22 | 1818,23 | 2318,58 | 3255,81 | 2818,15 | 2761,32 | 3289,23 | 3134,26 | 3725,27 | 1965,28 |
RNA isolation and cDNA synthesis
Investigator Kira
Motivation: in order to figure out if our cell lines do express telomerase, cell pellets were used for RNA isolation.
RNeasy Kit was used for RNA isolation. The cells were harvested and pellets washed 2x with PBS.
add RLT buffer
homogenize the lysate with syringe ans blunt needle
add 1 volume of 70% EtOH
centrifuge for 15s @ 10.000 rpm
add 700 ul Buffer RW1 and centrifuge again
add 500 ul Buffer RPE and centrifuge
add 500 ul Buffer RPE and centifuge for 2 min
place the column in 1.5 ml tube and add RNase free water (50 ul)
concentrations:
c(293)= 41, 68 ng/ul
c(a431) = 135, 48 ng/ul
Gel apparatus (chamber, tray and comb) for RNA purity gel has to be prepared a day ahead with : water, SDS and NaOH
cDNA synthesis
Investigator: Kira
The yielded DNA was used for cDNA synthesis.
QuantiTect Reverse Transcription (Qiagen) reaction was performed according to the manufacturer protocol.
DNA elimination reaction mix:
| components | a431/µl | 293/µl |
| 7xgDNA Wipeout Buffer | 2 | 2 |
| Template RNA | 1,6 | 2 |
| RNase-free H2O | 10,4 | 10 |
| Total volume | 14 | 14 |
Reverse-transcription reaction mix:
| components | 1xreaction | MM |
| Quantiscript Reverse Transcriptase | 1 | 3 |
| 5x Quantiscript RT buffer | 4 | 12 |
| RT primer Mix | 1 | 3 |
| DNA elimination reaction mix | 14 | - |
| Total volume | 20 | - |
incubation for 15 min @ 42C
incubation for 3 min @ 95C
128. labday 23.09.2010
Colony PCR of new clones from the last ligations
Investigator Achim
asdf
Contuniation: Cloning pSB1C3_Viral Brick 587KO-empty (B274/P542) into pSB1C3_VP2/3_Capins (B438/P514)
Investigator Patrick
3 Clones were picked in the morning and in each case inoculated 10 ml DYT. Clone 3 didnt grew fast enough. There were not enough bacteria in the eppi even after 8 ml were centrifuged. The mini-prep of clone 1 and 2 was performed according to the standrd protocol yielding the following concentrations:
Clone 1: 36,64 ng/µl
Clone 2: 7,8 ng/µl
Therefore only clone 1 was sent for sequencing (labelling: pat1, primer:VR2) and only this clone received a plasmid number: P569
No test-digestion was performed.
Because all medium was used to receive a sufficient amount of bacteria from clone 1 there was no cell suspension left to prepare a glycerol stock. Therefore again each clone was used to inoculated 10 ml DYT, following a mini-prep tomorrow.
Sequencing results:
Obviously there is something wrong with the backbone or the sequencing.
Mini-Prep and test digestion of pCerulean_Affibody_VP2/3, pSB1C3_Affibody_middlelinker_EGFP_His, pSB1C3_001_RC_InsRepCap_KpnIback_VP2-ko
Investigator: Jessica
Comment:
Glycerol stocks were prepared:
- B465 = pCerulean_ZEGFR:1907_VP23 clone1
- B466 = pCerulean_ZEGFR:1907_VP23 clone2
- B467 = pSB1C3_ZEGFR:1907_middlelinker_EGFP_His clone1
- B468 = pSB1C3_ZEGFR:1907_middlelinker_EGFP_His clone2
- B469 = pSB1C3_001_RC_InsRepCap_KpnIbackVP2-KO clone1
- B470 = pSB1C3_001_RC_InsRepCap_KpnIbackVP2-KO clone2
Mini-Prep was performed according to standard protocol:
- P563 = pCerulean_ZEGFR:1907_VP23 clone1 = 227,4 ng/µl
- P564 = pCerulean_ZEGFR:1907_VP23 clone2 = 337,2 ng/µl
- P565 = pSB1C3_ZEGFR:1907_middlelinker_EGFP_His clone1 = 252,7 ng/µl
- P566 = pSB1C3_ZEGFR:1907_middlelinker_EGFP_His clone2 = 209,5 ng/µl
- P567 = pSB1C3_001_RC_InsRepCap_KpnIback_VP1-KO clone1 = 304,7 ng/µl
- P568 = pSB1C3_001_RC_InsRepCap_KpnIback_VP1-KO clone2 = 383,1 ng/µl
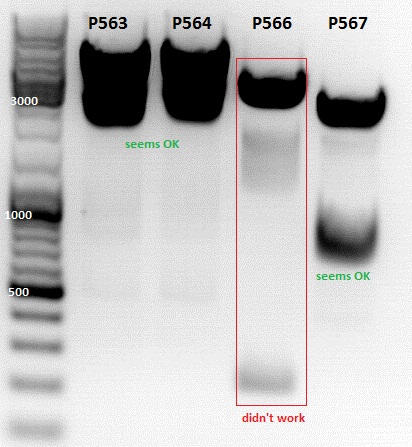
Test digestion of
- P563 = pCerulean_ZEGFR:1907_VP23 clone1
- P564 = pCerulean_ZEGFR:1907_VP23 clone2
- P565 = pSB1C3_ZEGFR:1907_middlelinker_EGFP_His clone1
- P566 = pSB1C3_ZEGFR:1907_middlelinker_EGFP_His clone2
| Components | P563-566 Volume/µL |
| DNA | 2 |
| BSA (10x) | 1 |
| Buffer no. 4 (10x) | 1 |
| PstI | 0,5 |
| XbaI | 0,5 |
| H2O | 5 |
| Total volume | 10 |
- P563 and P566 was sent for sequencing with primer VF2 and VF2/VR2
PCR for 587 Z34C ViralBricks
Investigators: Achim, Anna
- Plasmids used as template:
12_T4_1: c = 80,36 ng/µl
13_T4_2: c = 80,87 ng/µl
14_T4_2: c = 98,52 ng/µl
- Primer used:
| Template | Primer for | Primer rev |
| 12_T4_1 | RFC25 | O146 |
| 13_T4_2 | RFC25 | O148 |
| 14_T4_2 | O149 | RFC25 |
| Ingredients | Volume / µl |
| 5X Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| forward primer: see above | 2,5 |
| reverse primer: see above | 2,5 |
| DNA Template (1:100 dilution) | 1,0 |
| DMSO | 1,0 |
| Phusion Polymerase | 0,5 |
| H2O | 31,5 |
| Total volume | 50 |
PCR program:
| Cycles | Temperature / °C | Time / s |
| 98 | 60 | |
| 98 | 15 | |
| 25x | 68 | 25 |
| 72 | 10 | |
| 1x | 72 | 300 |
| Hold | 4°C |
Digestion of samples:
Restriction enzymes used: BamHI and PvuI.
Comment:
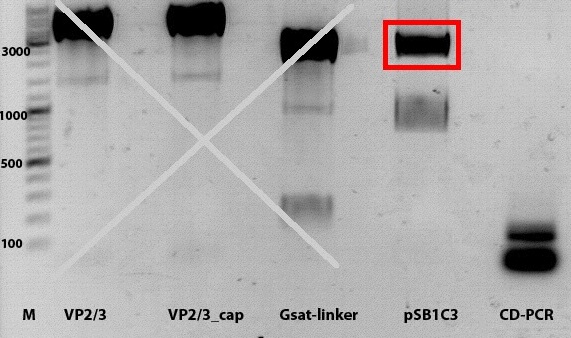
PCR of Vp1_ex, Vp2_ex and Vp3_ex and cloning into pSB1C3_001
Investigator: Anissa
- DNA sample P72 (pkex_Vp1), P73(pkex_Vp2) and P74(pkex_Vp3) were diluted 1:100
- used primers: O89 = Präfix_Vp1ex, O90= Präfix_Vp2ex, O91= Präfix_Vp2ex, O92= Suffix_Vp123ex, O120= Suffix_Vp123ex_reg
| Ingredients | Volume / µl |
| 5X Phusion HF buffer | 10 |
| 10 mM dNTP mix | 1 |
| forward primer: O152 | 2,5 |
| reverse primer: O188 | 2,5 |
| DNA Template | 2,0 |
| DMSO | 1 |
| Phusion Polymerase | 0,5 |
| H2O | 30,5 |
| Total volume | 50 |
PCR program:
| Cycles | Temperature / °C | Time / s |
| 1x | 98 | 60 |
| 25x | 98 | 15 |
| 25x | 60 | 25 |
| 25x | 72 | 55 |
| 1x | 72 | 5 |
| Hold | 4 |
Comment:The primer O120 was firstly given to the samples. It's a different primer for the Vp123_suffix, which binds to the regulatory-sequence. Because there was no knowledge, if the Vp_ex constucts have this sequence or not, O92 was also given to the samples.
Comment:Because gel-picture showed only DNA within the gel-bags, PCR was repeated.
Digestion of plasmid backbone:
Plasmid used: pSB1C3_001 (P320)
c (pSB1C3_001) = 408 ng/ µl
| Components | vector Volume/µL |
| DNA | 2,5 |
| BSA (10x) | 1,5 |
| Buffer no. 4 (10x) | 1,5 |
| SpeI | 1 |
| NgomIV | 1 |
| H2O | 7,5 |
| Total volume | 15 |
incubation @ 37 C for approx. 1,5 h
Comment:Strangely, PCR-products did not run into the gel, altough the PCR was repeated, after first PCR had two reverse primers in the sample. This could be because of a complex with the polymerase.However, ligation will be continued with the DNA within the bags.
</br>Digestion of PCR product:
| Components | PCR products Volume/µL |
| DNA | 20 |
| BSA (10x) | 3 |
| Buffer no. 4 (10x) | 3 |
| SpeI | 1 |
| NgomIV | 1 |
| H2O | 2 |
| Total volume | 30 |
Digestion was performed at 37 °C for 2 hours.
PCR-Purification:
- was peformed according the standard-protocol.
- c (P432) =-- ng/µl
Comment:THE WHOLE PCR WILL BE REPEATED ONCE AGAIN
RNA Purity Gel
Investigator: Kira
10x FA buffer: 200mM MOPS, 50mM Sodium Acetat, 10mM EDTA, RNase free water
1x FA running buffer: 10x FA buffer, 37% Formamid, RNAase free water
Agarose gel: Agarose 0,6 g; 10x FA Buffer 5ml; RNAse free water 45ml; 37% Formamid 0,9 ml
hTERT PCR Reaction
Investigator: Kira
| Ingredients | Volume / µl |
| 10x PCR buffer | 2 |
| 10 mM dNTP mix | 0,4 |
| 50mM MgCl2 | 0,6 |
| Primer 1 | 0,5 |
| Primer 2 | 0,5 |
| cDNA template | 1 |
| Taq Polymerase | 0,1 |
| H2O | 14,9 |
| Total volume | 20 |
PCR program:
| Cycles | Temperature / °C | Time |
| 1x | 94 | 3min |
| 30x | 94 | 45sec |
| 51 | 30 | |
| 72 | 1min 30sec | |
| 1x | 72 | 10 |
| Hold | 4 |
1% agarose gel:
Evaluation: The agarose gel reveals that all samples show telomerase activity but in different levels.
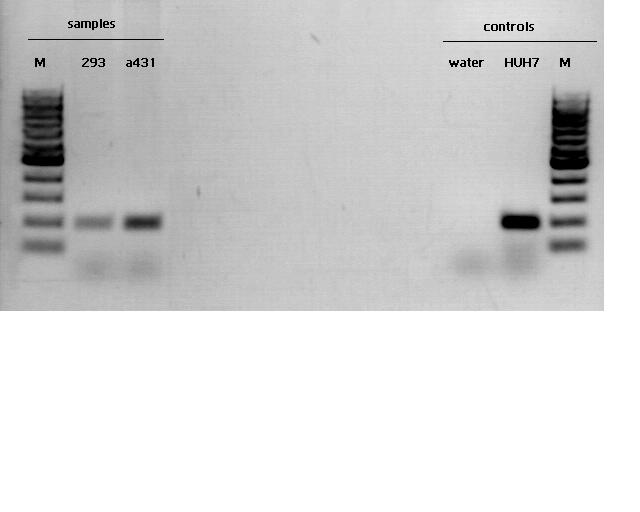
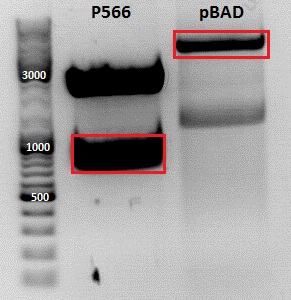
Cloning of pBAD_Affibody_middlelinker_EGFP_His
Investigator: Jessica
- Vector: name: pBAD_ OmpA His FluA LL Foki PGerrit
- Insert: name: pSB1C3_ZEGFR:1907_middlelinker_EGFP_His P566
- new vector name: pBAD_ZEGFR:1907_middlelinker_EGFP_His P
- buffer used: 4
- DNA concentration (vector): 97,0 ng/µl ; DNA concentration (insert): 209,5 µg/µl
| components | volume of pBAD_ OmpA His FluA LL Foki /µl | volume of pSB1C3_ZEGFR:1907_middlelinker_EGFP_His /µl |
| DNA | 10 | 6 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme XbaI | 0,8 | 0,8 |
| Enzyme AgeI | 0,8 | 0,8 |
| H2O | 4,4 | 8,4 |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 |
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 120 Volt
Loading plan for agarose gel:
Loading dye with SDS (6X), 3 µl
Marker used: GeneRuler ladder mix (Fermentas), 6 µl
| Marker | Sample PGerrit, 20µl | Sample 566, 20µl | |
|---|---|---|---|
| Lane | 1 | 3 | 5 |
| Fragment size | 4047 bp | 963 bp |
Gel extraction:
| pBAD | Affibody_middlelinker_EGFP_His | |
|---|---|---|
| DNA-concentration [ng/µl] | 8,3 | 10,2 |
T4 Ligation:
Volume vector: 5,06 µl
Volume insert: 2,94 µl
Transformation:
XL1B cells were used.
Picking clones of pSB1C3_CMV_VP123_inscap
Investigator: Anna
Three clones were picked (10 ml DYT, 10 µl Cm).
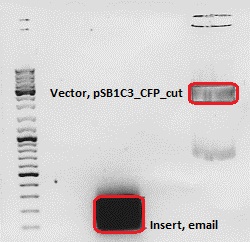
Cloning of ***** into pSB1C3_CFP
Investigator: Chris W.
- Vector: name: pSB1C3_CFP P51,2
- Insert: name: email Hybrid Oligo (O184&O185)
- new vector name: pSB1C3_CMV_email
- buffer used: 4
- DNA concentration (vector): 151,0 ng/µl ; DNA concentration (insert): 95,3 µg/µl
| components | volume of pSB1C3_CFP /µl | volume of email/µl |
| DNA | 6 | 15 |
| BSA (10x) | 2 | 2 |
| Buffer 4 (10x) | 2 | 2 |
| Enzyme I | NgoMIV 1 | NgoMIV 1 |
| Enzyme II | SpeI 1 | SpeI 1 |
| H2O | 8,4 | - |
| Total volume (e.g. 15,20,25,30 µl) | 20 | 20 |
0,5 g Agarose, 50 ml TAE (1%), 3 µl GELRED , at 120 Volt, running time:45
Loading plan for agarose gel:
Loading dye with SDS (6X), 4 µl
Marker used: GeneRuler ladder mix (Fermentas), 3 µl
| Marker | Sample Insert, 20µl | Sample Vector, 20µl | |
|---|---|---|---|
| Lane | 1 | 3 | 5 |
| Fragment size | 70 bp | 2070 bp |
Gel extraction:
| pSB1C3_CFP_cut | ||
|---|---|---|
| DNA-concentration [ng/µl] | 7,8 | 27,4 |
T4 Ligation:
Volume vector: 8,74 µl
Volume insert: 0,26 µl
Transformation:
XL1B cells were used.
Repetition: Cloning of P5_TATAless downstream of pSB1C3_001_RC_insrepcap_KpnI_back and P5 upstream of pSB1C3_001_RC_insrepcap_KpnI_back
Investigator: Stefan
Digestion of the constructs:
- P432 = pSB1C3_001_RC_insrepcap_KpnI_back c=344,5 ng/µl
- P432 = pSB1C3_001_RC_insrepcap_KpnI_back c=344,5 ng/µl
- P180 = pSB1C3_pTAV2 (P5)clone 1 c=108,8 ng/µl
- P184 = pSB1C3_pAAV_RC (P5TATAless)clone 3 c=176,9 ng/µl
| Components | V P432up /µL | VP180up /µL | V P432down /µL | V P184down /µL |
| DNA | 4,5 | 14 | 4,5 | 11 |
| BSA (10x) | 2 | 2 | 2 | 2 |
| Buffer no. 4 (10x) | 2 | 2 | 2 | 2 |
| Enzyme 1 | XbaI 1 | SpeI 1 | SpeI 1 | XbaI 1 |
| Enzyme 2 | EcoRI 1 | EcoRI 1 | PstI 1 | PstI 1 |
| H2O | 9,5 | 9,5 | - | 3 |
| Total volume | 20 | 20 | 20 | 20 |
Results:
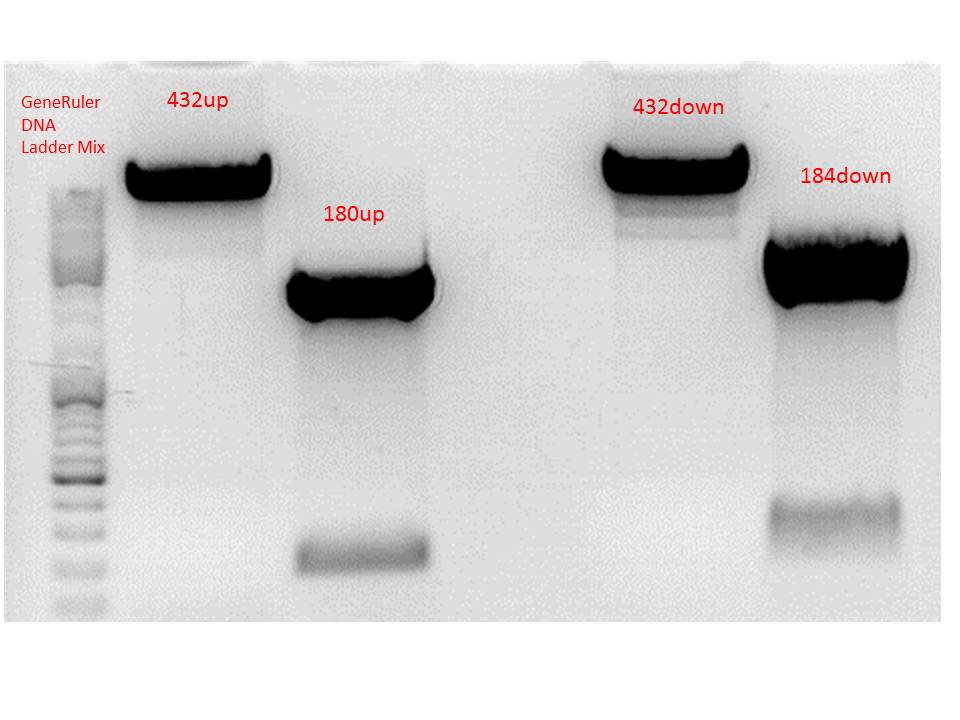
Gel-Extraction:
Gel-Ex was performed according to protocol. The following concentrations were measured:
- 432up c= 25,73 ng/µl
- 180up c= 9,57 ng/µl
- 432down c= 27,34 ng/µl
- 184down c= 17,80 ng/µl
Ligation:
The following DNA mixes were used for ligation:
- 432up V= 6,3 µl
- 180up V= 1,7 µl
- 432down V= 6,93 µl
- 184down V= 1,07 µl
Transformation:
The transformation was performed according to standard protocol using BL21 cells. They were plated on agar plates containing chloramphenicol and stored in 37°C room overnight.
129. labday 24.09.2010
Contuniation: Cloning pSB1C3_Viral Brick 587KO-empty (B274/P542) into pSB1C3_VP2/3_Capins (B438/P514)
Investigator Patrick
Because of the sequencing results of clone 1 a mini-prep was performed only with clone 2 and 3 yielding the following concentrations:
pSB1C3_VP2/3_Capins_587KO-empty clone 2: 195 ng/µl
pSB1C3_VP2/3_Capins_587KO-empty clone 3: 224,74 ng/µl
Testdigestion:
pSB1C3_VP2/3_Capins_587KO-empty clone 2: 3 µl Buffer 4, 0,5 µl BamHI, 0,5 µl PvuII, 22 µl DNA, 4 µl H2O
pSB1C3_VP2/3_Capins_587KO-empty clone 3: 23 µl Buffer 4, 0,5 µl BamHI, 0,5 µl PvuII, 20 µl DNA, 6 µl H2O
Because somebody switched the 37°C thermoblock off i dont know the exact digestion time. Therefore i switched the thermoblock on again and waited 1 hour. I used that much DNA because the desired insert is 48 bp long.
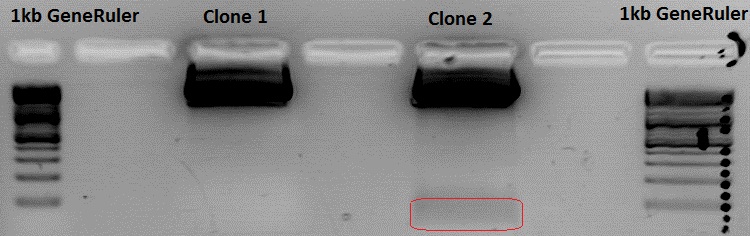
Clone 3 was sent for sequencing.
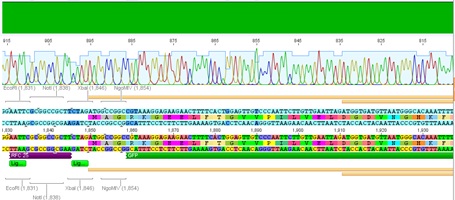
Sequencing evaluation of VP2/3_Gsat-linker and VP2/3_cap_Gsat-linker
Investigator Kira
Comment: pSB1C3_VP2/3_cap_Gsat-linker as well as pSB1C3_VP2/3_Gsat-linker can be used for further experiments.
New CD primer ordered
Investigator Stefan
Comment: CD reverse primer was incorrect. A new primer was ordered.
 "
"