Apoptosis Notebook
Contents
8-02-2010
8-03-2010
Some test text in bold
We created following tests:
- test4
- test5
8-04-2010
Example of a table
| header 1
| header 2
| header 3
|
| row 1, cell 1
| row 1, cell 2
| row 1, cell 3
|
| row 2, cell 1
| row 2, cell 2
| row 2, cell 3
|
8-05-2010
this too is a table:
| H2Oddes
| 10,3 µl
|
| RE10 + Buffer H
| 2,0 µl
|
| acetylated BSA
| 0,2 µl
|
| DNA
| 6,0 µl
|
table with 3 cells
| apple | banana | peaches
|
| green | yellow | red
|
8-06-2010
text
8-07-2010
text
8-08-2010
test
test
8-09-2010
text Knallroter Text
farbnummern für farbige schrift: http://html.nicole-wellinger.ch/hilfen/farbenverzeichnis.html
test grüner text
8-10-2010
Transforming competent cells
- eGFP Biobrick: BBa_I714891 SDY_eGFP (Kanamycin)
- TEV recogn N Degron SF3 = pDS7 (Ampicillin)
- TEV p14 recogn = 190-6 (Ampicillin)
-> Protocol: (3 Transformation)
- We added 2 µl DNA
- We plated out 200 µl
Plasmid Isolation
- CMV-Promoter Biobrick: BBa_J52034
-> Protocol:(4 Plasmid extraction from cells)
- Prepared overnight culture, measured concentration of DNA
-> Poor results -> thrown away
8-11-2010
New Plasmid Extraction
- CMV-Promoter Biobrick: BBa_J52034
-> Protocol: (4 Plasmid extraction from cells)
- Plasmid concentration: 143ng/µl
Prepared overnight culture of eGFP BBa_I714891
- 3 ml LB-Media + 4 µl Kanamycin
- Inoculated iangeimpft) with 1 colony of BBa_I714891 -> 37°C
Prepared overnight culture of 190-6 and pDS7 and eGFP (BBa_I714891) in falcons
- for 190-6 and pDS7: 10µl Ampicillin + 10 ml LB-Media + colony of plate
- for eGFP: 13,3 µl Kanamycin + 10 ml LB-Media + 1 colony of plate
Restriction digest (Restriktionsverdau) of CMV-Promoter BBa_J52034 with EcoRI and PstI
| H2Oddest, sterile
| 10,3 µl
|
| RE10 + Buffer H
| 2,0 µl
|
| acetylated BSA (18ng/µl)
| 0,2 µl
|
| DNA (0,143µg/µl)
| 6,0 µl
|
-> mixed
- plus: EcoRI (10µg/µl): 0,5 µl resp. PstI (10µg/µl): 0,5 µl
- incubated at room temperature from 12:10 to 15:00, 1 hour at 37°C, 2 hours at 60°C
- frozen at -20°C
Prepare new/fresh overnight culture of CMV-Promoter Biobrick: BBa_J52034
- 1 ml of "old" culture + 3 ml LB-Media + 4 µl Kanamycin -> 37°C
8-12-2010
- plasmid extraction of pDS7 (458ng/µl), eGFP (55ng/µl), 190-6 (193ng/µl)
-> Protocol: (4 Plasmid extraktion from cells)
- Restriction digest with EcoRI and PstI in buffer H (for testing DNA is correct)
10µg DNA: pDS7 (2µl), eGFP (15µl), 190-6 (10µl)
-> Protocol: (5 Restriction digest)
- Colony for Plasmidextraction (CMV (Kanamycin), eGFP (Kanamycin), pDS7 (Ampicillin), 190-6 (Ampicillin)) plated
- PhiC31o plated on Ampicillin-Agar, stored at 37°C
- 50% Glycerol made (for the glycerolstock PhiC31o)
8-13-2010
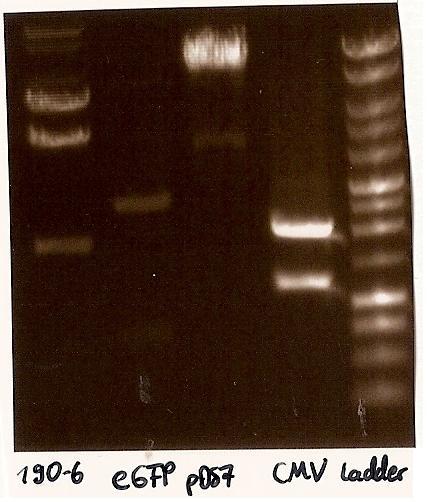 Gelfoto from the EcoR1 and Pst1 Restrictiondigest of 190-6, eGFP, pDS7 and CMV - CMV (BBa_J52034) from 10.8.2010 inoculated into LB medium with ampicillin, as falsly inoculated in Kanamycin
- Agarosegelelectrophoresis with the digestions (CMV, eGFP, pDS7, 190-6), 125V for 30 minutes and then for 20 minutes;
expected DNA bands: 190-6 (4840bp, 1903bp), pDS7 (8027bp, 6bp), CMV (654 bp (Insert), 2079bp (Plasmid)), eGFP (720bp (Insert), 2750bp (Plasmid))
- Correct DNA bands for 190-6 (~4800bp, ~1900bp, ~6700bp (undigested plasmid)) and eGFP (~2000bp (Plasmid), ~750 bp (Insert)); CMV probably not digested (two bands; one probably normal, one supercoiled) and pDS7 not clear
- Restriction digest from CMV (EcoR1, Pst1; 6µl DNA, buffer H) and pDS7 (EcoR1, Spe1; 2µl DNA, buffer B)
Expected DNA bands: CMV see above, pDS7 (3647bp, 3369bp, 1011bp, 6bp)
-> Protocol (5 Restriction digest)
- Agarosegelelectorphoresis for 30 minutes, 150V
- false DNA bands CMV (~1200 bp, ~2000 bp) and pDS7 (~8000bp two bands, ~1100 bp); required to isolate a new colony for these two Plasmidextractions
- Plated the colony from CMV (BBa_J52034) for Plasmidextraction (Ampicillin), as falsly plated on Kanamycin
8-14-2010
weekend
8-15-2010
weekend
8-16-2010
- transfer 1 ml PhiC31o culture to new LB medium + Amp, 37°C
- pick up CMV and pDS7 colonies from plates and transfer to LB medium+Amp, 37°C
- plasmid extraction of PhiC310 ->27,5ng/µl DNA and second plasmid extraction of PhiC310 (i. o. to get more DNA); first eluation-step with first eluation-extraction
-> 60ng/µl DNA
->Protocol (4 Plasmid extraktion from cells)
- restriction digest of PhiC310 with EcoR1 and Spe1
| H2Oddest, sterile
| 0 µl
|
| Buffer B
| 2,0 µl
|
| BSA (1:10)
| 2 µl
|
| DNA (0,06µg/µl)
| 15,0 µl
|
| EcoR1
| 0,5 µl
|
| Spe1
| 0,5µl
|
restriction digest in the thermo cycler (program "Verdau")
->Protocol (5 Restriction digest)
- Primer handling after arrival
->Protocol (9 Handling primers)
- 10mM dNTP mix made from 100 mM dATP, dGTP, dCTP, dTTP by taking 100µl of each and adding 600µl H 2 O
-PCR of the tet inducible CMV minimal promotor out of prevTRE (=PCR 1 with Primer 1 and 2) and SV40PA out of pcDNA3 (=PCR 6 with Primer 11 and 12)
Mixture:
|
| pcDNA3 (0,6 µg/µl)
| pTRERev (0,15µg/µl)
|
| Primer
| 2*2,5µl (P1+P2)
| " (P11+P12)
|
| 300ng template
| 0,5µl
| 2µl
|
| 10x Buffer Pfu
| 5µl
| "
|
| dNTP Mix
| 1µl
| "
|
| Pfu Polymerase (3u/µl)
| 0,5µl
| "
|
| H2O
| 40,5µl
| 39µl
|
| summ
| 52,5µl
| 52,5µl
|
Programme:
| Denaturation
| 95°C
| 2min
|
| 30 times:
| Denaturation
| 95°C
| 1min
|
|
| Annealing
| 45°C
| 30sec
|
|
| Extension
| 73°C
| 2min
|
| Final Extension
| 73°C
| 5min
|
| Soak (end)
| 12°C
| infinite
|
->Protocol (10 PCR with Pfu)
- Glycerolstock of the colony of PhiC31o for the plasmidextraction
| bacterial culture
| 800µl
|
| Glycerol (50%)
| 500µl
|
8-17-2010
text
8-18-2010
Next iGEM meeting at 6pm!
8-19-2010
Workshop with Tanya on Wiki/Web page at 6pm
8-20-2010
text
8-21-2010
text
8-22-2010
text
|


![]()
![]()







![]()
![]()

![]()
![]()
![]()

 "
"
