Team:Peking/Modeling
From 2010.igem.org
Evamonlight (Talk | contribs) (→References) |
Evamonlight (Talk | contribs) |
||
| Line 18: | Line 18: | ||
</html> | </html> | ||
=Introduction= | =Introduction= | ||
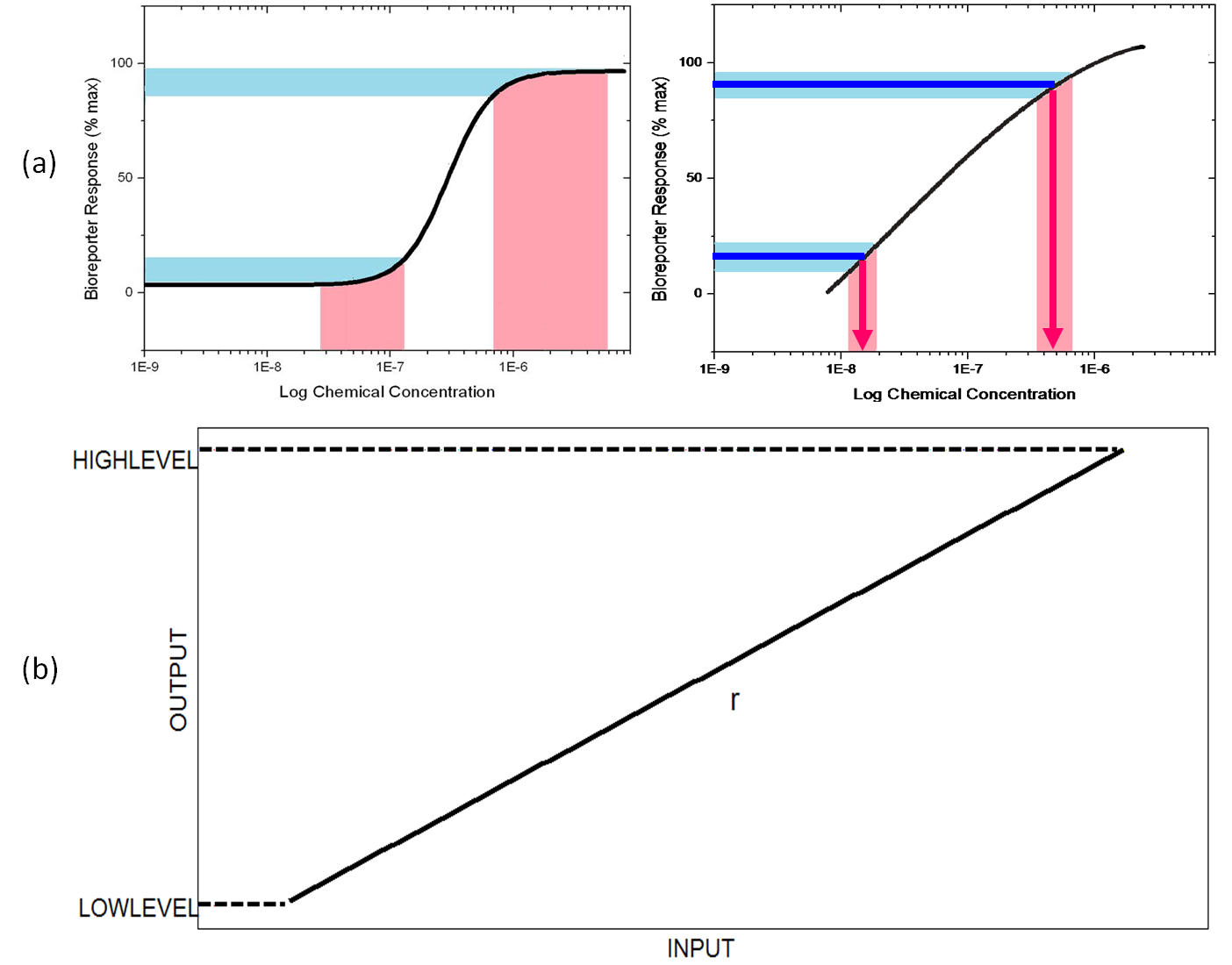
| + | The biological systems are full of noise, and the response of bioreporters represents a Hill Function. Noise will be significantly amplified and the output detected will be invalid at the ends of dose response curve.(Figure 1(a)) Therefore, The detection is limited at a narrow range. In order to solve the problem, we seek to transform the Hill Function curve to a straight line. As Figure 1(a) shows, the noise is reducted, and the sensitivity and detectable-range-characteristic of biosensors will be significantly improved. | ||
<html> | <html> | ||
| - | We adopted the process of Reverse Engineering which in our work means to enumerate all possible network topologies and analyzed whether they | + | We adopted the process of Reverse Engineering which in our work means to enumerate all possible network topologies and analyzed whether they function, thus getting the right topology. In details, |
<br> | <br> | ||
| - | Here we | + | Here we name our target functionality as Input-Output Alignment(IOA). In order to define IOA precisely for need of calculation, we considered most important characters of IOA and adopted Pearson Correlation Coefficient r to represent Input-Output Linear Relationship in the overall search work ( when r>0.99 we consider the network topology having the IOA function ), and also, regulated two levels for the initial and ultimate output concentration for the second character – the output range in further search work.(Figure 1) |
<br> | <br> | ||
<br></html> | <br></html> | ||
| - | [[Image: | + | [[Image:Intro2.png|center|600px]] |
<br> | <br> | ||
<br> | <br> | ||
Revision as of 08:27, 27 October 2010

Modeling Home
Introduction
The biological systems are full of noise, and the response of bioreporters represents a Hill Function. Noise will be significantly amplified and the output detected will be invalid at the ends of dose response curve.(Figure 1(a)) Therefore, The detection is limited at a narrow range. In order to solve the problem, we seek to transform the Hill Function curve to a straight line. As Figure 1(a) shows, the noise is reducted, and the sensitivity and detectable-range-characteristic of biosensors will be significantly improved.
We adopted the process of Reverse Engineering which in our work means to enumerate all possible network topologies and analyzed whether they function, thus getting the right topology. In details,
Here we name our target functionality as Input-Output Alignment(IOA). In order to define IOA precisely for need of calculation, we considered most important characters of IOA and adopted Pearson Correlation Coefficient r to represent Input-Output Linear Relationship in the overall search work ( when r>0.99 we consider the network topology having the IOA function ), and also, regulated two levels for the initial and ultimate output concentration for the second character – the output range in further search work.(Figure 1)
Figure 1 Factors for selection of IOA network topologies. r is the Pearson Correlation Coefficient and the output range is HIGHLEVEL minus LOWLEVEL.
Calculating Process
In this part, we will demostrate our calculating process in three sections--Network enumeration, Equations set up and network topologies’ analysis.
== Learn more ==
Analyses and Results
Here we analyzed the results we obtained and found the final right topology.
== Learn more ==
Advanced Model
When the range of input is very large, the IOA networks no longer meet out needs and so we tried to find one semilog-linear topology in this section.
== Learn more ==
References
1 Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K. & Walter, P. (2002)Molecular Biology of the Cell (Garland, New York).
2 Nicolas E. Buchler, Ulrich Gerland, Terence Hwa, Nonlinear protein degradation and the function of genetic circuits[J], PNAS, 2005, 102(27):9559-9564.
3 John D. Helmann, Barry T. Ballard, Christopher T. Walsh, The MerR Metalloregulatory Protein Binds Mercuric Ion as a Tricoordinate, Metal-Bridged Dimer[J], Science, 1998, 247:946-948.
4 Diana. M, Ralston and Thomas V. O Halloran, Ultrasensitivity and heavy-metal selectivity of the allosterically modulated MerR transcription complex[J], PNAS, 1990, 87:3846-3850.
5 Uri Alon, An Introduction to Systems Biology—Design Principles of Biological Circuits[M], Chapman&Hall/CRC,2007.
6 Wickner, S., Maurizi, M. R. & Gottesman, S.Posttranslational Quality Control: Folding, Refolding, and Degrading Proteins[J], Science, 1999, 286:1888–1893.
7 W. Ma, A. Trusina, H. El-Samad, W.A. Lim, C. Tang. Defining network topologies that can achieve biochemical adaptation[J], Cell, 2009, 138:760–773.
 "
"