Team:Monash Australia/Modelling
From 2010.igem.org
(→Model 1) |
(→Model 1) |
||
| Line 64: | Line 64: | ||
----- | ----- | ||
<H4> Graphs and results</H4> | <H4> Graphs and results</H4> | ||
| - | [[Image:graph1|300px|thumb|left|EThylene Production at different steady state concentration]] | + | [[Image:graph1.jpg|300px|thumb|left|EThylene Production at different steady state concentration]] |
== <H2>Model 2</H2>== | == <H2>Model 2</H2>== | ||
Revision as of 07:37, 27 October 2010

|
|
|||
Contents |
Introduction
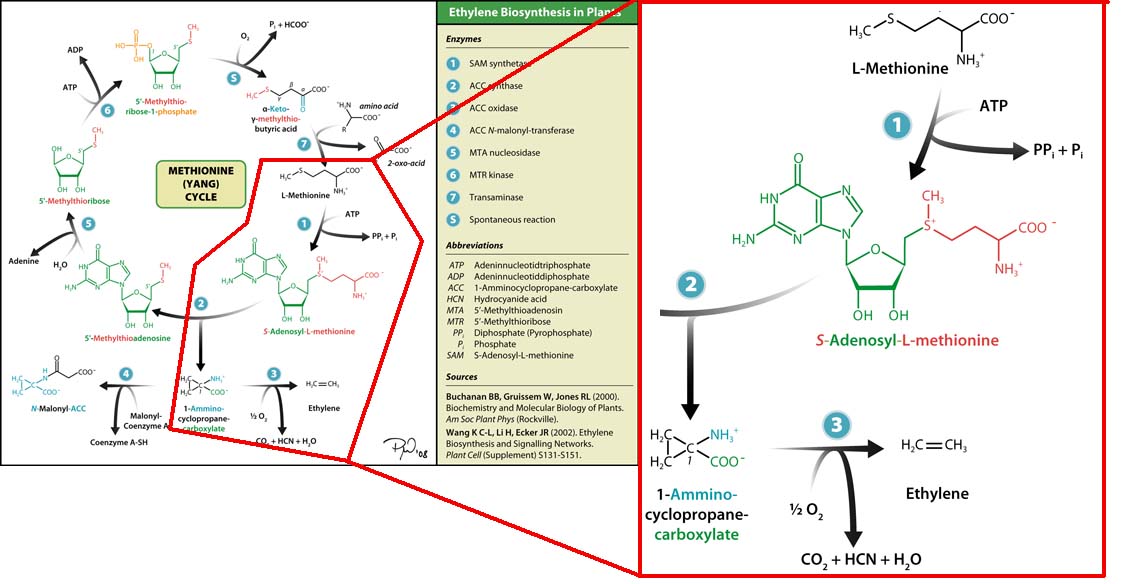
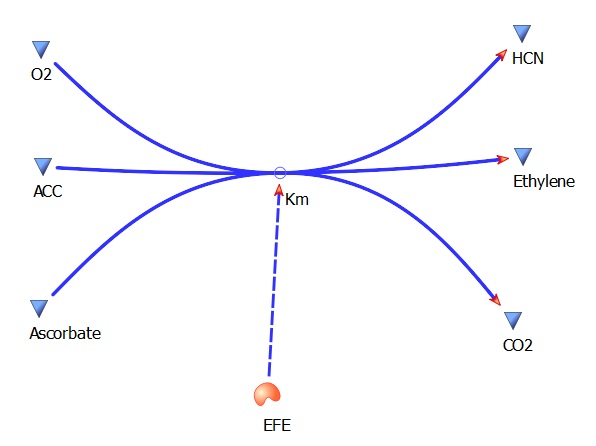
The three key enzymes we require are highlighted in the image, SAM synthase, ACC synthase and ACC oxidase. SAM synthase converts methionine into S-Adenosyl-L-Methionine (SAM), using ATP for an adensoyl group. The second step involves ACC synthase, which cleaves the amino butyrate from SAM, releasing 1-aminocyclopropane-1-carboxylic acid (ACC). Released ACC is then processed by ACC Oxidase which converts ACC to ethylene by cleaving the carboxylic acid off as carbon dioxide and its neighboring carbon with the amino group as cyanide gas. By using such a system to produce ethyene gas we can potentially reduce costs involved with current production methods by reducing temperature requirements by 30 fold.
Kinetic modelling of the ethylene generator
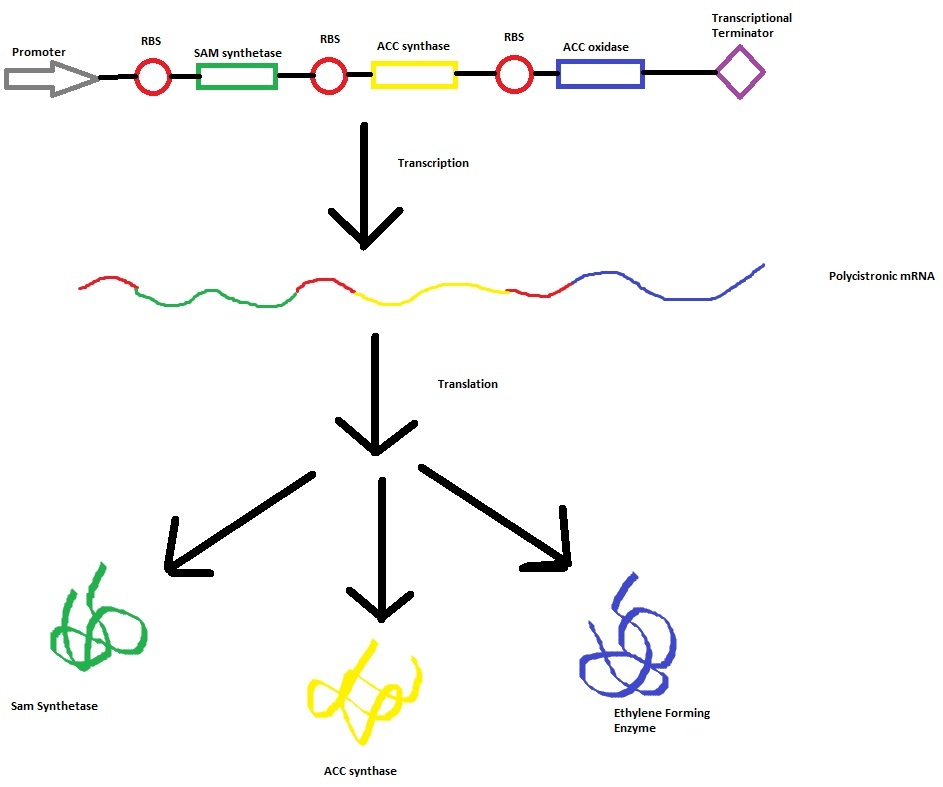
The program tinkercell was used to model the synthesis of ethylene in our e.coli cell. Where, in addition to the qualitative representation (figure 1) the program allowed, simulation of the model could be exercised inorder to measure the flux of metabolites – with respect to time - in the system given the input of values pertaining to substrate/enzyme concentration, and reaction rates were inputted. In addition to experimental data, with a model, theoretical data can be obtained using the model to extrapolate quantitative data corresponding to mRNA, protein and ethylene outputs under set parameters and assumptions in silico. As a result, using the given theoretical model outputs as a benchmark, manipulations to the parameters and assumptions set can be made in vitro/in vivo in order to achieve higher outputs in future studies of the 'ethylene generator'.
Aims of the Kinetic Modelling
-To determine the maximum output of ethylene -To estimate HCN levels and assess potential damage to cell
-Start with a simple model and progress to a more complex one. This allows us to more accurately predict ethylene production
Model 1
Introduction to Model 1
This model was designed to be the starting point of the our kinetic modelling. Therefore it was decided to begin with a realitivly simple construct.This uncomplicated system could then be used to obtain an ethylene output. This design comprised only of three enzymes and lacked the transcription and translation stages. The enzymes invloved were SAM synthetase, ACC synthase and ACC oxidase or EFE (ethylene forming enzyme). These enzymes were presnt at fixed concentrations (10, 100, 1000 and 3000 uM) and in the 1:1:1 ratio.. This ensured that the model was plain yet ultimately obtained a resonable yet simple ethylene output. Therefore with fixed enzyme concentrations the rate at which ethylene is produced depends on the Vmax of the specific enzymes.
This specific rate is defined by the Michaelis-Menton Equation
kcat*[substrate]*[enzyme]
Rate = -------------------------
km + [substrate]
Assumptions for Model 1
- A 1:1:1 steady state of enzymes- No production inhibition
-Substrates (ATP, L-Methionine etc) kept at a constant value due to homeostatic mecahnisms in E.coli
- The best value (km, kcat etc) was always chosen to ensure maxiumum yield of ethylene
Parameters and Values used
Met concentration - 150 uMATP concentration - 9600 uM
O2 concentration - 442 uM
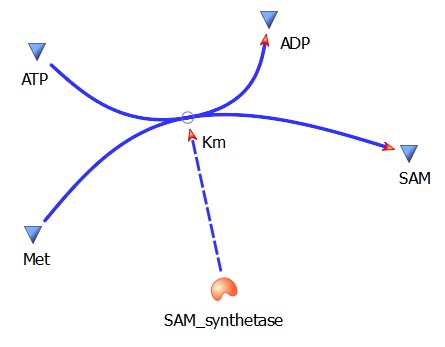
SAMsynth Km(Met)- 92 uM
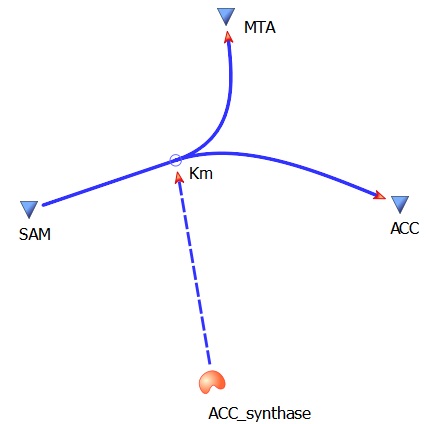
ACCsynth Km(AdoMet)- 37 uM
EFE Km(ACC)- 12 uM
SAMsynth turnover rate (kcat)- 1.5 per second
ACCsynth turnover rate (kcat)- 18 per second
EFE turnover rate (kcat) - 5.9 per second
Graphs and results
Model 2
Model 2 explores the more complicated version of our kinetic modelling. It aims to incorporate the transcription and translation rates of the cell into the final design. This calculation begins to become more difficult due to problomatic degredation,transcription and translation rates. However these values where obtained from the [http://www.ebi.ac.uk/biomodels-main/BIOMD0000000012 Elowitz repressilator model]. Of the many sources browsed it was determined that this source seemed to be the most accurate and hence its values were used. Therefore the transcription and translation rates of mRNA and proteins constantly changes the final concentration of the three enzymes. This changing concentration of enzymes then produces ethylene at a more reliable rate. Therefore this model is designed to produce a more accurate representation of the ethylene producing biological pathway
Elowitz Degradation Values
mRNA degradation rate - 0.00577 mRNA per second Enzyme degradation rate - 0.001155 enzymes per second R0011 PoPs or transcription rate - 0.5 mRNA per second Translation rate -0.117 enzymes per secondGraphs and results
more to come
Construct --> mRNA --> enzymes (SAMsynth, ACCS, EFE)
Met --> SAM (AdoMet)
SAM --> ACC
ACC --> Ethylene
More to come soon!
 "
"