Team:USTC Software/Features
From 2010.igem.org
| Line 1: | Line 1: | ||
{{Team:USTC_Software/Header}} | {{Team:USTC_Software/Header}} | ||
| - | { | + | {| |
|- | |- | ||
| + | |style="padding: 20px 20px 20px 20px;"| | ||
==Overall project== | ==Overall project== | ||
The ultimate goal of USTC_Software 2010 team is to promote synthetic biology throughout the world. To attract more people who do not have biology background to be interested in this area, we plan to develop a modeling-and-simulation game specially designed for synthetic biology. Users are taught to learn the basic knowledge in the area via constructing their genetic circuits as input to our software and try to understand the system behavior as output. It is the basic function module. We also plan to develop a rating system to grade users' design for given tasks, such as construction of logic gates or oscillators. Though more functions are expected, we focus on the basic module, modeling-and-simulaton, at the first of our project. | The ultimate goal of USTC_Software 2010 team is to promote synthetic biology throughout the world. To attract more people who do not have biology background to be interested in this area, we plan to develop a modeling-and-simulation game specially designed for synthetic biology. Users are taught to learn the basic knowledge in the area via constructing their genetic circuits as input to our software and try to understand the system behavior as output. It is the basic function module. We also plan to develop a rating system to grade users' design for given tasks, such as construction of logic gates or oscillators. Though more functions are expected, we focus on the basic module, modeling-and-simulaton, at the first of our project. | ||
Revision as of 05:52, 23 October 2010
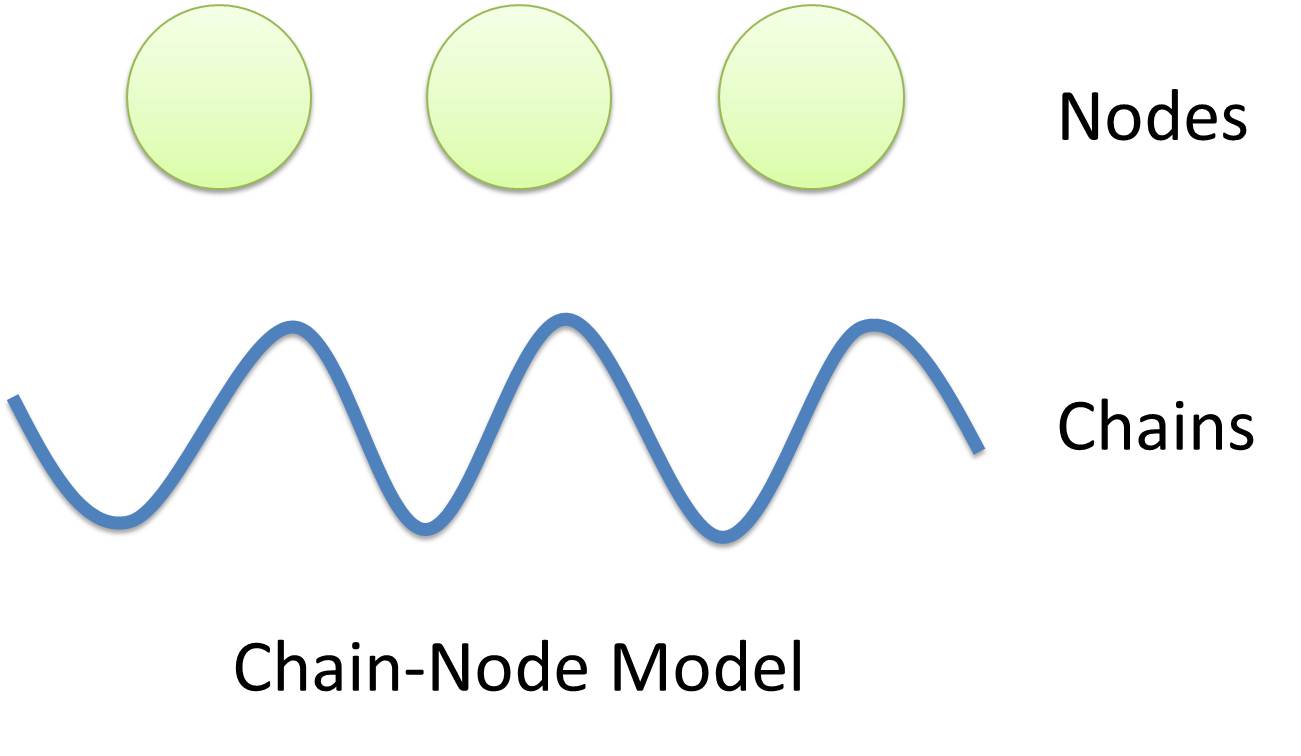
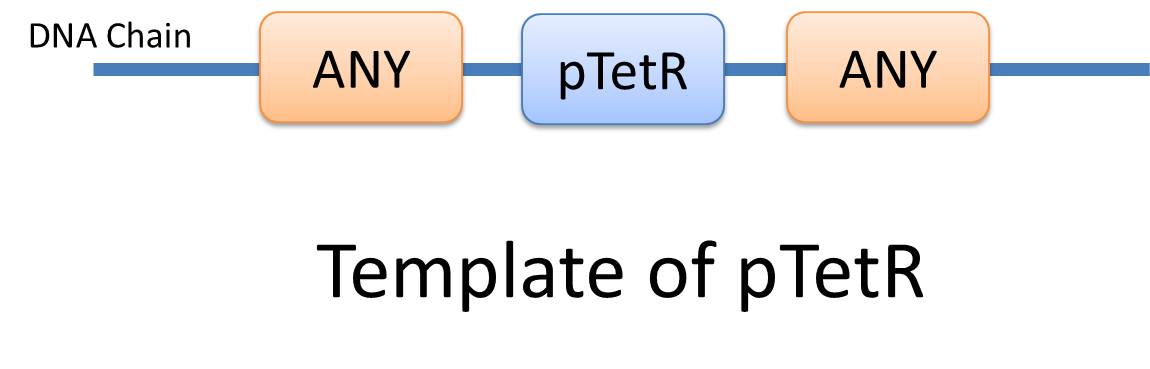
Overall projectThe ultimate goal of USTC_Software 2010 team is to promote synthetic biology throughout the world. To attract more people who do not have biology background to be interested in this area, we plan to develop a modeling-and-simulation game specially designed for synthetic biology. Users are taught to learn the basic knowledge in the area via constructing their genetic circuits as input to our software and try to understand the system behavior as output. It is the basic function module. We also plan to develop a rating system to grade users' design for given tasks, such as construction of logic gates or oscillators. Though more functions are expected, we focus on the basic module, modeling-and-simulaton, at the first of our project. In practice, many CAD (Computer-Aided Design) tools, such as TinkerCell and Synbioss, have been developed to model and simulate biological system and give the system behavior as guides. However, they all need users to provide details of the system network, such as the activation and repression of transcription and translation reactions in genetic regulatory network. It is so difficult for even professionals to construct a detailed network of a complex system only depending on their minds. In this sense, modeling through CAD tools will not reduce the work of modeling: users are actually required to model manually and input their model in details. However, since our software is developed for non-biological background users, it is unrealistic to expect them to model their design manually. To solve this problem, the USTC_Software 2010 team attempts to take synthetic biology modeling a step further by introducing new methods for automatic modeling of biological systems. Just as automation implies, users are only required to submit their assembling of parts for our program to discover and generate the biological model automatically. Being the first-ever team trying to develop a synthetic biology automatic modeling tool, we emphasize on genetic regulatory network at the first year and develop our software tool, iGame, which will assist in the design of genetic function modules for biological systems in synthetic biology. We believe this will greatly relieve users from handling complex interactions of species in biological system. Many novel and revolutionary concepts are proposed during our development. The first is our Chain-Node model for complex structure with multiple chains bound together. It is necessary for automation since behaviors of species (such as how it react with other species) must be determined by their structures instead of their names: it is impossible to construct a universal name-based reaction database applying for different systems. The next is Template. A species with a certain structure is a template species, and a reaction occuring between several template species is a template reaction. The introduction of Template makes it possible to describe a group of reactions with same structure-determined reaction mechanism. Finally, based on our Chain-Node and Template concept, we propose an XML-based Standard Biological Part Automatic Modeling Database Language (MoDeL for abbreviation) to fully characterize Species and Reaction in templates with clear definitions of elements and attributes in XML fashion. It is a database language for next generation when automatic modeling is widely used throughout the world. Though we have no enough time to achieve our ultimate goal completely, we have successfully developed a MoDeL-based, automatic modeling and simulation software. Our program falls into 3 major components. The first component is user interface. Users could give their assembling of parts by drag-and-pull function and setup initial conditions as well. System behavior as output will also be shown there. The second component is database written in MoDeL, which is the kernel of our automatic modeling idea. And the last is our core program designed to support MoDeL language. It functions as a driver: comleting system network based on users' input and data stored in our database to give dynamic analysis as output. First look at our features
Bring Biological Modeling to the Next Level
Modeling with Templates
Automatic Modeling Database Language
| ||||||||||||||||
 "
"